News
| 11.2024 | Our work "A Generic Residue Numbering Scheme for the GAIN Domain of adhesion G protein-coupled receptors" has just been accepted for publication in Nature Communications. The GAIN domain is a hotspot for pathological mutations. However, the low primary sequence conservation of GAIN domains has thus far hindered the knowledge transfer across different GAIN domains in human receptors as well as species orthologs. To overcome this obstacle, we developed the GAIN-GRN scheme in collaboration with the David Gloriam Laboratory at Copenhagen University that enables researchers to generate hypothesis and rationalize experiments related to GAIN domain function and pathology. | |
| 10.2024 | Article accepted in Nature Reviews Drug Discovery "Functional dynamics of G protein coupled receptors reveal new routes for drug discovery" by Vittorio Limongelli, Paolo Conflitti, Ed Lyman, Mark Sansom, Hugo Guitérrez-de-Terán, Paolo Carloni, Bertie Ansell, Shuguang Yuan, Patrick Barth, Anne Robinson, Peter Hildebrand, Matthew Eddy, Scott Prosser, Stephan Grzesiek, David Gloriam, Christopher G. Tate, and Vittorio Limongelli. | |
| 06.2024 | Our article: "mechanistic insights into G-protein coupling with an agonist-bound G-protein-coupled receptor" hast been accepted at the journal nature structural & molecular biology. Here to the publication |
|
| 02.2024 | Our collaborative work with Yiorgo Skiniotis lab has just been accepted by NATURE you'll find the preprint here |
|
| 06.2023 | Press release: Novel TRENDS Article on aGPCRs out! 7TM domain structures of adhesion GPCRs: what's new and what's missing? Seufert, F., Chung, Y. K., Hildebrand, P. W., Langenhan, T. Trends in Biochemical Sciences (2023) Here to the publication |
|
| 05.2023 | Pressemitteilung: Das Leipziger Universitätsmagazin: "Neue digitale Werkzeuge, um das Beste aus beiden Welten miteinander zu verbinden" Wie zwei Top-Wissenschaftler die Wirkungsweise von Rezeptoren erforschen Hier zum Artikel des Leipziger Universitätsmagazin |
 |
| 03.2023 | We proudly announce the release of our new website MutationExplorer. | 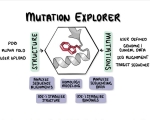
|
| 05.2022 | Our update on the MDsrv, a collaborative work with Gerik Scheuermann and Daniel Wiegreffe from the Informatics Institute has just been accepted to be published in the upcoming webserver issue of Nucleic Acids Research:
Michelle Kampfrath, René Staritzbichler, Guillermo Pérez Hernández, Alexander S. Rose, Johanna K.S. Tiemann, Gerik Scheuermann, Daniel Wiegreffe, Peter W. Hildebrand MDsrv -- visual sharing and analysis of molecular dynamics simulations |

|
| 02.2022 | Our collaborative work with the Smith and the Huster lab, from the Instiut of Medical Physics and Biophysics of Leipzig University on membranee dynamics has just been accepted at Nat. Commun. | |
| 07.2021 | The winner of the 2021 Doctoral Award of the Leipzig Medical Faculty is Johanna Tiemann with her dissertation "Interactive Analysis of Protein Structure and Dynamics". We wish her continued success in her scientific career. |  |
| 30.05.2021 | Jugend forscht: Nikola hat mit dem Voronoia Projekt, an dem er seit 2 Jahren bei uns unter der Betreuung von René Staritzbichler im Rahmen einer besonderen Lernleistung mitarbeitet, den Bundeswettbewerb von Jugend Forscht in Chemie gewonnen. Herzlichen Glückwunsch Nikola! Zum Presseartikel der LVZ, Presseartikel des Stern, Interview des MDR oder zur Website von Jugend forscht. |
|
| 18.05.2021 | The webservice Voronoia has been updated and accepted to be published in the upcoming webserver issue of nucleic acid research. | 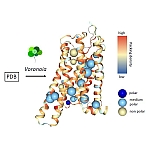 |
| 15.02.2021 | Our collaborative work with the Langenhan and the Sauer lab, from Leipzig and Würzburg University on a GPCR activation has just been published... | |
| 18.12.2020 | New updated version of the voronoia server is online now | 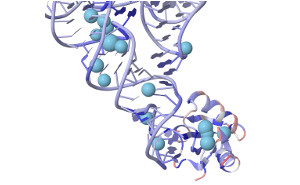 |
| 01.09.2020 | Please find our new publication in PNAS, which is a contribution to Brian Kobilkas work: Analysis of β2AR-Gs and β2AR-Gi complex formation by NMR spectroscopy. doi: 10.1073/pnas.2009786117 PNAS More publications here... |
|
| 21.1.2020 | Two PhD positions are available at the Hildilab in the newly funded collaborative research centre on GPCR dynamics!
The job posting is closed. |
|
| 08.11.2019 | Our TIBS article 'Bringing Molecular Dynamics Simulation Data into View.' on the cover. | 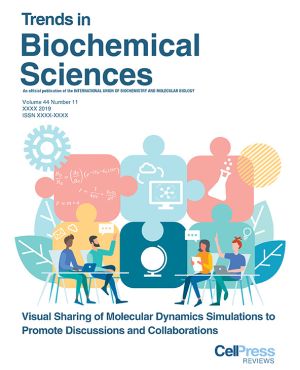 |
| 17.09.2019 | We are happy to announce this years workshop on structural modeling and MD simulations which will be organized together with the Jens Meiler Laboratory from Vanderbilt University. Find the accordant link here: Modeling protein mutations with Rosetta | 
|
| 02.09.2019 | Our project “In silico GPCR: 'A computational microscope to determine receptor – G protein coupling specificity and functional selectivity'” at the Berlin Institut of Health, funded by the Stiftung Charite was renewed until March 31, 2022. This establishes another 2 years of funding for the Berlin laboratory of BIH Visiting Fellow Brian Kobilka from Standford University. | |
| 10.07.2019 | Our TIBS Opinion into 'Bringing Molecular Dynamics Simulation Data int\o View.' is going online. | |
| 07.06.2019 | Please find our new publication in CELL, which is a contribution to Brian Kobilkas work on structural intermediates that covern receptor-G protein complex formation. In this context, please find a recent press release of the Berlin Institute of Health. | 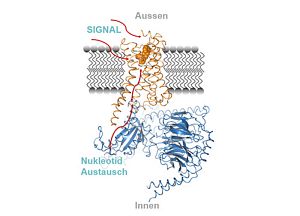 |
| 28.05.2019 | A promising SFB evaluation. | |
| 24.05.2019 | The new website is online. |  |
