Start search
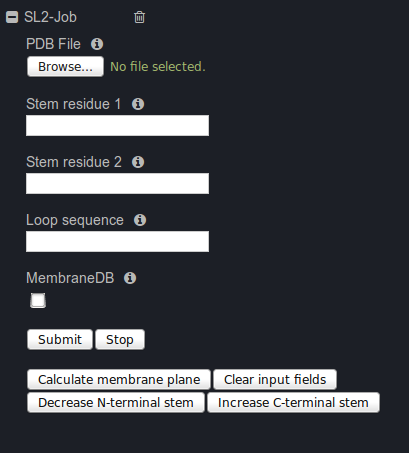
To start a new SL2 job you have to provide a coordinate file of a protein structure in PDB format.
The two stem residues flanking a missing segment or a segment that should be remodeled have to be
typed in a form, or selected from the NGL visualization by mouse selection. To clear the stem residues
from the form, use the 'Clear input fields' button, afterwards, they can be filled by mouse selection again.
Finally, the amino acids sequence of the missing segment has to be provided. To search for loops only
originating from membrane proteins check the MembraneDB check-box, otherwise the full database will be
searched. Additionally, the membrane can be calculated by applying informations from
TMDET [3] and some further processing.
Depending on fragment length, a fragment search may take up to 30 seconds. When the job is
finished, the results are displayed automatically. To stop a running job press the 'Stop' button.
Please be aware that the protein structure that should be used for the loop search has to be uploaded via the
file upload dialog of the SL2 job formular (not by drag and drop). To visualize additional molecules that are not used to search loops
drag the file into the viewer. If additional molecules are loaded, be aware of selecting only stem residues
of the molecule loaded by the 'SL2 job formular'.
Modify search
If no suitable loop was found, one possible solution is to extend the loop search in N- or C- terminal direction by pushing the "Decrease N-terminal stem" or "Increase C-terminal stem" button. The stem residue will be changed by one into the chosen direction
and the search sequence will be updated automatically. To modify the search parameters manually change the form and press
submit. The result table will be updated after the search is completed. To start a completely new job, please reload the web-site. Old results will be lost.
|
 |