Binding-Pocket Interactions of Four EGFR Inhibitors¶
For this notebook, we use mdciao to visualize the binding-pocket interactions of four Epidermal Growth Factor Receptor (EGFR) inhibitors. EGFR is an important drug target with implications in cancer and inflammation (Wikipedia). It is a transmembrane protein with an extracellular receptor domain and an intracellular kinase domain.
The molecular dynamics (MD) data used here was generated by slightly modifying the notebook
which is part of the impressive TeachOpenCADD collection, made available as teaching platform for computer-aided drug design by the Volkamer Lab at Charité Univeristätsmedizin Berlin.
The four inhibitors and structures are chosen from the following RCSB entries:
The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 (PDB ID 7VRE)
EGFR kinase domain complexed with compound 20a (PDB ID 3W32)
Crystal Structure of EGFR(L858R/T790M/C797S) in complex with CH7233163 (PDB ID 6LUB)
Please see the references at the bottom of the notebook for more information.
[1]:
import mdciao
import os
import matplotlib
import nglview
from glob import glob
Consensus labeler object for KLIFS nomenclature¶
Since it will be used more than once, it is better to have it instantiated only once and reused many times. The only thing we need is the UniProt Accession Code of the EGFR, P00533.
[2]:
KLIFS = mdciao.nomenclature.LabelerKLIFS("P00533")
No local file ./KLIFS_P00533.xlsx found, checking online in
https://klifs.net/api/kinase_ID?kinase_name=P00533 ...done!
Please cite the following reference to the KLIF structural database:
* Kanev et al, (2021) KLIFS: an overhaul after the first 5 years of supporting kinase research
Nucleic Acids Research 49, D562--D569
https://doi.org/10.1093/NAR/GKAA895
For more information, call mdciao.nomenclature.references()
Please cite the following reference to the UniProt Knowledgebase:
* Bateman et al, (2021) UniProt: the universal protein knowledgebase in 2021
Nucleic Acids Research 49, D480--D489
https://doi.org/10.1093/NAR/GKAA1100
For more information, call mdciao.nomenclature.references()
done without 404, continuing.
Download example data¶
[3]:
if not os.path.exists("example_kinases"):
mdciao.examples.fetch_example_data("EGFR");
Guess molecular fragments¶
[4]:
for pdb in sorted(glob("example_kinases/*.pdb")):
print(pdb)
mdciao.fragments.get_fragments(pdb)
print()
example_kinases/topology.3POZ.pdb
Auto-detected fragments with method 'lig_resSeq+'
fragment 0 with 317 AAs GLN701 ( 0) - LEU1017 (316 ) (0)
fragment 1 with 1 AAs 03P1 ( 317) - 03P1 (317 ) (1)
example_kinases/topology.3W32.pdb
Auto-detected fragments with method 'lig_resSeq+'
fragment 0 with 317 AAs GLN701 ( 0) - LEU1017 (316 ) (0)
fragment 1 with 1 AAs W321 ( 317) - W321 (317 ) (1)
example_kinases/topology.6LUB.pdb
Auto-detected fragments with method 'lig_resSeq+'
fragment 0 with 323 AAs GLY696 ( 0) - ILE1018 (322 ) (0)
fragment 1 with 1 AAs EUX1 ( 323) - EUX1 (323 ) (1)
example_kinases/topology.7VRE.pdb
Auto-detected fragments with method 'lig_resSeq+'
fragment 0 with 323 AAs GLY696 ( 0) - ILE1018 (322 ) (0)
fragment 1 with 1 AAs 7VH1 ( 323) - 7VH1 (323 ) (1)
All three setups share the equivalent topology of kinase (fragment 0) and ligand (fragment 1):
from PDB ID
3POZligandP31from PDB ID
3W32ligandW321from PDB ID
6LUBligandEUX1from PDB ID
7VREligand7VH1
For labelling purposes, create a mapping between PDB IDs and ligand names:
[5]:
pdb2lig = {"6LUB" : "EUX1",
"7VRE" : "7VH1",
"3W32" : "W321",
"3POZ" : "P31"}
Compute the ligand-kinase interactions for the four inhibitors¶
[6]:
binding_pocket = {}
for pdb in sorted(glob("example_kinases/*.pdb")):
key = os.path.basename(pdb).split(".")[1]
key="%s@%s"%(pdb2lig[key], key)
xtc = pdb.replace(".pdb",".xtc").replace("topology","trajectory")
binding_pocket[key]=mdciao.cli.interface(xtc,
pdb,
fragment_names=["EGFR", "ligand"],
KLIFS_uniprotAC=KLIFS,
ctc_control=1.0,
frag_idxs_group_1=[0],
frag_idxs_group_2=[1],
ctc_cutoff_Ang=4, interface_cutoff_Ang=None,
accept_guess=True, figures=False, no_disk=True)
Will compute contact frequencies for trajectories:
example_kinases/trajectory.3POZ.xtc
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 317 AAs GLN701 ( 0) - LEU1017 (316 ) (0)
fragment 1 with 1 AAs 03P1 ( 317) - 03P1 (317 ) (1)
KLIFS-labels align best with fragments: [0] (first-last: GLN701-LEU1017).
These are the KLIFS fragments mapped onto your topology:
I with 3 AAs LYS716@I.1 ( 15) - LEU718@I.3 (17 ) (I)
g.l with 6 AAs GLY719@g.l.4 ( 18) - GLY724@g.l.9 (23 ) (g.l)
II with 4 AAs THR725@II.10 ( 24) - LYS728@II.13 (27 ) (II)
III with 6 AAs VAL742@III.14 ( 41) - LEU747@III.19 (46 ) (III)
αC with 11 AAs GLU758@αC.20 ( 57) - SER768@αC.30 (67 ) (αC)
b.l with 7 AAs VAL769@b.l.31 ( 68) - ARG776@b.l.37 (75 ) (b.l) resSeq jumps
IV with 4 AAs LEU777@IV.38 ( 76) - ILE780@IV.41 (79 ) (IV)
V with 3 AAs GLN787@V.42 ( 86) - ILE789@V.44 (88 ) (V)
GK with 1 AAs THR790@GK.45 ( 89) - THR790@GK.45 (89 ) (GK)
hinge with 3 AAs GLN791@hinge.46 ( 90) - MET793@hinge.48 (92 ) (hinge)
linker with 4 AAs PRO794@linker.49 ( 93) - CYS797@linker.52 (96 ) (linker)
αD with 7 AAs LEU798@αD.53 ( 97) - GLU804@αD.59 (103 ) (αD)
αE with 5 AAs TYR827@αE.60 ( 126) - ARG831@αE.64 (130 ) (αE)
VI with 3 AAs ARG832@VI.65 ( 131) - VAL834@VI.67 (133 ) (VI)
c.l with 8 AAs HIS835@c.l.68 ( 134) - ASN842@c.l.75 (141 ) (c.l)
VII with 3 AAs VAL843@VII.76 ( 142) - VAL845@VII.78 (144 ) (VII)
VIII with 1 AAs ILE853@VIII.79 ( 152) - ILE853@VIII.79 (152 ) (VIII)
xDFG with 4 AAs THR854@xDFG.80 ( 153) - GLY857@xDFG.83 (156 ) (xDFG)
a.l with 2 AAs LEU858@a.l.84 ( 157) - ALA859@a.l.85 (158 ) (a.l)
Computing distances in the interface between fragments
0
and
1
0%| | 0/1 [00:00<?, ?it/s]
Streaming example_kinases/trajectory.3POZ.xtc (nr. 0) with stride 1 in chunks of 10000 frames. Now at chunk nr 0, frames so far 535
100%|█████████████████████████████████████████████████████████████████████| 1/1 [00:00<00:00, 1.52it/s]
These 28 contacts capture 20.38 (~99%) of the total frequency 20.50 (over 317 input contacts)
As orientation value, the first 21 ctcs already capture 90.0% of 20.50.
The 21-th contact has a frequency of 0.44
freq label residue idxs sum
0 1.00 C775@b.l.36 - P31@ligand 74 317 1.00
1 1.00 K745@III.17 - P31@ligand 44 317 2.00
2 1.00 M793@hinge.48 - P31@ligand 92 317 3.00
3 1.00 T854@xDFG.80 - P31@ligand 153 317 4.00
4 1.00 Q791@hinge.46 - P31@ligand 90 317 5.00
5 1.00 T790@GK.45 - P31@ligand 89 317 5.99
6 1.00 L844@VII.77 - P31@ligand 143 317 6.99
7 0.99 D855@xDFG.81 - P31@ligand 154 317 7.98
8 0.99 A743@III.15 - P31@ligand 42 317 8.97
9 0.98 F856@xDFG.82 - P31@ligand 155 317 9.96
10 0.96 L788@V.43 - P31@ligand 87 317 10.92
11 0.96 L777@IV.38 - P31@ligand 76 317 11.88
12 0.92 L792@hinge.47 - P31@ligand 91 317 12.80
13 0.92 R776@b.l.37 - P31@ligand 75 317 13.72
14 0.92 V726@II.11 - P31@ligand 25 317 14.64
15 0.91 M766@αC.28 - P31@ligand 65 317 15.55
16 0.81 L718@I.3 - P31@ligand 17 317 16.36
17 0.72 S720@g.l.5 - P31@ligand 19 317 17.08
18 0.71 R841@c.l.74 - P31@ligand 140 317 17.79
19 0.52 F997@EGFR - P31@ligand 296 317 18.31
20 0.44 I744@III.16 - P31@ligand 43 317 18.75
21 0.36 G796@linker.51 - P31@ligand 95 317 19.11
22 0.34 G719@g.l.4 - P31@ligand 18 317 19.45
23 0.26 L858@a.l.84 - P31@ligand 157 317 19.71
24 0.19 C797@linker.52 - P31@ligand 96 317 19.91
25 0.17 N842@c.l.75 - P31@ligand 141 317 20.08
26 0.16 G721@g.l.6 - P31@ligand 20 317 20.24
27 0.14 I789@V.44 - P31@ligand 88 317 20.38
label freq
0 C775@b.l.36 1.00
1 K745@III.17 1.00
2 M793@hinge.48 1.00
3 T854@xDFG.80 1.00
4 Q791@hinge.46 1.00
5 T790@GK.45 1.00
6 L844@VII.77 1.00
7 D855@xDFG.81 0.99
8 A743@III.15 0.99
9 F856@xDFG.82 0.98
10 L788@V.43 0.96
11 L777@IV.38 0.96
12 L792@hinge.47 0.92
13 R776@b.l.37 0.92
14 V726@II.11 0.92
15 M766@αC.28 0.91
16 L718@I.3 0.81
17 S720@g.l.5 0.72
18 R841@c.l.74 0.71
19 F997@EGFR 0.52
20 I744@III.16 0.44
21 G796@linker.51 0.36
22 G719@g.l.4 0.34
23 L858@a.l.84 0.26
24 C797@linker.52 0.19
25 N842@c.l.75 0.17
26 G721@g.l.6 0.16
27 I789@V.44 0.14
label freq
0 P31@ligand 20.38
Will compute contact frequencies for trajectories:
example_kinases/trajectory.3W32.xtc
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 317 AAs GLN701 ( 0) - LEU1017 (316 ) (0)
fragment 1 with 1 AAs W321 ( 317) - W321 (317 ) (1)
KLIFS-labels align best with fragments: [0] (first-last: GLN701-LEU1017).
These are the KLIFS fragments mapped onto your topology:
I with 3 AAs LYS716@I.1 ( 15) - LEU718@I.3 (17 ) (I)
g.l with 6 AAs GLY719@g.l.4 ( 18) - GLY724@g.l.9 (23 ) (g.l)
II with 4 AAs THR725@II.10 ( 24) - LYS728@II.13 (27 ) (II)
III with 6 AAs VAL742@III.14 ( 41) - LEU747@III.19 (46 ) (III)
αC with 11 AAs GLU758@αC.20 ( 57) - SER768@αC.30 (67 ) (αC)
b.l with 7 AAs VAL769@b.l.31 ( 68) - ARG776@b.l.37 (75 ) (b.l) resSeq jumps
IV with 4 AAs LEU777@IV.38 ( 76) - ILE780@IV.41 (79 ) (IV)
V with 3 AAs GLN787@V.42 ( 86) - ILE789@V.44 (88 ) (V)
GK with 1 AAs THR790@GK.45 ( 89) - THR790@GK.45 (89 ) (GK)
hinge with 3 AAs GLN791@hinge.46 ( 90) - MET793@hinge.48 (92 ) (hinge)
linker with 4 AAs PRO794@linker.49 ( 93) - CYS797@linker.52 (96 ) (linker)
αD with 7 AAs LEU798@αD.53 ( 97) - GLU804@αD.59 (103 ) (αD)
αE with 5 AAs TYR827@αE.60 ( 126) - ARG831@αE.64 (130 ) (αE)
VI with 3 AAs ARG832@VI.65 ( 131) - VAL834@VI.67 (133 ) (VI)
c.l with 8 AAs HIS835@c.l.68 ( 134) - ASN842@c.l.75 (141 ) (c.l)
VII with 3 AAs VAL843@VII.76 ( 142) - VAL845@VII.78 (144 ) (VII)
VIII with 1 AAs ILE853@VIII.79 ( 152) - ILE853@VIII.79 (152 ) (VIII)
xDFG with 4 AAs THR854@xDFG.80 ( 153) - GLY857@xDFG.83 (156 ) (xDFG)
a.l with 2 AAs LEU858@a.l.84 ( 157) - ALA859@a.l.85 (158 ) (a.l)
Computing distances in the interface between fragments
0
and
1
0%| | 0/1 [00:00<?, ?it/s]
Streaming example_kinases/trajectory.3W32.xtc (nr. 0) with stride 1 in chunks of 10000 frames. Now at chunk nr 0, frames so far 558
100%|█████████████████████████████████████████████████████████████████████| 1/1 [00:00<00:00, 1.44it/s]
These 28 contacts capture 22.01 (~100%) of the total frequency 22.08 (over 317 input contacts)
As orientation value, the first 22 ctcs already capture 90.0% of 22.08.
The 22-th contact has a frequency of 0.51
freq label residue idxs sum
0 1.00 C775@b.l.36 - W321@ligand 74 317 1.00
1 1.00 F856@xDFG.82 - W321@ligand 155 317 2.00
2 1.00 D855@xDFG.81 - W321@ligand 154 317 3.00
3 1.00 T854@xDFG.80 - W321@ligand 153 317 4.00
4 1.00 T790@GK.45 - W321@ligand 89 317 5.00
5 1.00 M793@hinge.48 - W321@ligand 92 317 6.00
6 1.00 K745@III.17 - W321@ligand 44 317 7.00
7 1.00 L788@V.43 - W321@ligand 87 317 7.99
8 0.99 A743@III.15 - W321@ligand 42 317 8.99
9 0.99 Q791@hinge.46 - W321@ligand 90 317 9.98
10 0.99 L844@VII.77 - W321@ligand 143 317 10.96
11 0.99 R776@b.l.37 - W321@ligand 75 317 11.95
12 0.97 M766@αC.28 - W321@ligand 65 317 12.91
13 0.96 G719@g.l.4 - W321@ligand 18 317 13.87
14 0.95 L777@IV.38 - W321@ligand 76 317 14.83
15 0.92 L792@hinge.47 - W321@ligand 91 317 15.75
16 0.92 V726@II.11 - W321@ligand 25 317 16.67
17 0.92 S720@g.l.5 - W321@ligand 19 317 17.58
18 0.85 L718@I.3 - W321@ligand 17 317 18.43
19 0.78 G796@linker.51 - W321@ligand 95 317 19.22
20 0.54 L1001@EGFR - W321@ligand 300 317 19.75
21 0.51 G721@g.l.6 - W321@ligand 20 317 20.27
22 0.45 I744@III.16 - W321@ligand 43 317 20.71
23 0.39 C797@linker.52 - W321@ligand 96 317 21.11
24 0.32 L858@a.l.84 - W321@ligand 157 317 21.42
25 0.23 I789@V.44 - W321@ligand 88 317 21.66
26 0.20 R841@c.l.74 - W321@ligand 140 317 21.85
27 0.15 F997@EGFR - W321@ligand 296 317 22.01
label freq
0 C775@b.l.36 1.00
1 F856@xDFG.82 1.00
2 D855@xDFG.81 1.00
3 T854@xDFG.80 1.00
4 T790@GK.45 1.00
5 M793@hinge.48 1.00
6 K745@III.17 1.00
7 L788@V.43 1.00
8 A743@III.15 0.99
9 Q791@hinge.46 0.99
10 L844@VII.77 0.99
11 R776@b.l.37 0.99
12 M766@αC.28 0.97
13 G719@g.l.4 0.96
14 L777@IV.38 0.95
15 L792@hinge.47 0.92
16 V726@II.11 0.92
17 S720@g.l.5 0.92
18 L718@I.3 0.85
19 G796@linker.51 0.78
20 L1001@EGFR 0.54
21 G721@g.l.6 0.51
22 I744@III.16 0.45
23 C797@linker.52 0.39
24 L858@a.l.84 0.32
25 I789@V.44 0.23
26 R841@c.l.74 0.20
27 F997@EGFR 0.15
label freq
0 W321@ligand 22.01
Will compute contact frequencies for trajectories:
example_kinases/trajectory.6LUB.xtc
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 323 AAs GLY696 ( 0) - ILE1018 (322 ) (0)
fragment 1 with 1 AAs EUX1 ( 323) - EUX1 (323 ) (1)
KLIFS-labels align best with fragments: [0] (first-last: GLY696-ILE1018).
These are the KLIFS fragments mapped onto your topology:
I with 3 AAs LYS716@I.1 ( 20) - LEU718@I.3 (22 ) (I)
g.l with 6 AAs GLY719@g.l.4 ( 23) - GLY724@g.l.9 (28 ) (g.l)
II with 4 AAs THR725@II.10 ( 29) - LYS728@II.13 (32 ) (II)
III with 6 AAs VAL742@III.14 ( 46) - LEU747@III.19 (51 ) (III)
αC with 11 AAs GLU758@αC.20 ( 62) - SER768@αC.30 (72 ) (αC)
b.l with 7 AAs VAL769@b.l.31 ( 73) - ARG776@b.l.37 (80 ) (b.l) resSeq jumps
IV with 4 AAs LEU777@IV.38 ( 81) - ILE780@IV.41 (84 ) (IV)
V with 3 AAs GLN787@V.42 ( 91) - ILE789@V.44 (93 ) (V)
GK with 1 AAs MET790@GK.45 ( 94) - MET790@GK.45 (94 ) (GK)
hinge with 3 AAs GLN791@hinge.46 ( 95) - MET793@hinge.48 (97 ) (hinge)
linker with 4 AAs PRO794@linker.49 ( 98) - SER797@linker.52 (101 ) (linker)
αD with 7 AAs LEU798@αD.53 ( 102) - GLU804@αD.59 (108 ) (αD)
αE with 5 AAs TYR827@αE.60 ( 131) - ARG831@αE.64 (135 ) (αE)
VI with 3 AAs ARG832@VI.65 ( 136) - VAL834@VI.67 (138 ) (VI)
c.l with 8 AAs HIS835@c.l.68 ( 139) - ASN842@c.l.75 (146 ) (c.l)
VII with 3 AAs VAL843@VII.76 ( 147) - VAL845@VII.78 (149 ) (VII)
VIII with 1 AAs ILE853@VIII.79 ( 157) - ILE853@VIII.79 (157 ) (VIII)
xDFG with 4 AAs THR854@xDFG.80 ( 158) - GLY857@xDFG.83 (161 ) (xDFG)
a.l with 2 AAs ARG858@a.l.84 ( 162) - ALA859@a.l.85 (163 ) (a.l)
Computing distances in the interface between fragments
0
and
1
0%| | 0/1 [00:00<?, ?it/s]
Streaming example_kinases/trajectory.6LUB.xtc (nr. 0) with stride 1 in chunks of 10000 frames. Now at chunk nr 0, frames so far 500
100%|█████████████████████████████████████████████████████████████████████| 1/1 [00:00<00:00, 1.36it/s]
These 24 contacts capture 17.65 (~99%) of the total frequency 17.84 (over 323 input contacts)
As orientation value, the first 18 ctcs already capture 90.0% of 17.84.
The 18-th contact has a frequency of 0.61
freq label residue idxs sum
0 1.00 M793@hinge.48 - EUX1@ligand 97 323 1.00
1 1.00 L718@I.3 - EUX1@ligand 22 323 2.00
2 1.00 Q791@hinge.46 - EUX1@ligand 95 323 3.00
3 1.00 P794@linker.49 - EUX1@ligand 98 323 4.00
4 1.00 M790@GK.45 - EUX1@ligand 94 323 4.99
5 1.00 L792@hinge.47 - EUX1@ligand 96 323 5.99
6 0.99 V726@II.11 - EUX1@ligand 30 323 6.98
7 0.97 G796@linker.51 - EUX1@ligand 100 323 7.95
8 0.97 G724@g.l.9 - EUX1@ligand 28 323 8.92
9 0.96 L844@VII.77 - EUX1@ligand 148 323 9.87
10 0.94 K745@III.17 - EUX1@ligand 49 323 10.82
11 0.93 A743@III.15 - EUX1@ligand 47 323 11.74
12 0.83 T725@II.10 - EUX1@ligand 29 323 12.57
13 0.82 T854@xDFG.80 - EUX1@ligand 158 323 13.39
14 0.78 G721@g.l.6 - EUX1@ligand 25 323 14.17
15 0.71 K728@II.13 - EUX1@ligand 32 323 14.87
16 0.69 S797@linker.52 - EUX1@ligand 101 323 15.56
17 0.61 G719@g.l.4 - EUX1@ligand 23 323 16.17
18 0.51 C775@b.l.36 - EUX1@ligand 79 323 16.68
19 0.30 F795@linker.50 - EUX1@ligand 99 323 16.99
20 0.28 D855@xDFG.81 - EUX1@ligand 159 323 17.27
21 0.15 D800@αD.55 - EUX1@ligand 104 323 17.42
22 0.12 S720@g.l.5 - EUX1@ligand 24 323 17.54
23 0.11 R841@c.l.74 - EUX1@ligand 145 323 17.65
label freq
0 M793@hinge.48 1.00
1 L718@I.3 1.00
2 Q791@hinge.46 1.00
3 P794@linker.49 1.00
4 M790@GK.45 1.00
5 L792@hinge.47 1.00
6 V726@II.11 0.99
7 G796@linker.51 0.97
8 G724@g.l.9 0.97
9 L844@VII.77 0.96
10 K745@III.17 0.94
11 A743@III.15 0.93
12 T725@II.10 0.83
13 T854@xDFG.80 0.82
14 G721@g.l.6 0.78
15 K728@II.13 0.71
16 S797@linker.52 0.69
17 G719@g.l.4 0.61
18 C775@b.l.36 0.51
19 F795@linker.50 0.30
20 D855@xDFG.81 0.28
21 D800@αD.55 0.15
22 S720@g.l.5 0.12
23 R841@c.l.74 0.11
label freq
0 EUX1@ligand 17.65
Will compute contact frequencies for trajectories:
example_kinases/trajectory.7VRE.xtc
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 323 AAs GLY696 ( 0) - ILE1018 (322 ) (0)
fragment 1 with 1 AAs 7VH1 ( 323) - 7VH1 (323 ) (1)
KLIFS-labels align best with fragments: [0] (first-last: GLY696-ILE1018).
These are the KLIFS fragments mapped onto your topology:
I with 3 AAs LYS716@I.1 ( 20) - LEU718@I.3 (22 ) (I)
g.l with 6 AAs GLY719@g.l.4 ( 23) - GLY724@g.l.9 (28 ) (g.l)
II with 4 AAs THR725@II.10 ( 29) - LYS728@II.13 (32 ) (II)
III with 6 AAs VAL742@III.14 ( 46) - LEU747@III.19 (51 ) (III)
αC with 11 AAs GLU758@αC.20 ( 62) - SER768@αC.30 (72 ) (αC)
b.l with 7 AAs VAL769@b.l.31 ( 73) - ARG776@b.l.37 (80 ) (b.l) resSeq jumps
IV with 4 AAs LEU777@IV.38 ( 81) - ILE780@IV.41 (84 ) (IV)
V with 3 AAs GLN787@V.42 ( 91) - ILE789@V.44 (93 ) (V)
GK with 1 AAs MET790@GK.45 ( 94) - MET790@GK.45 (94 ) (GK)
hinge with 3 AAs GLN791@hinge.46 ( 95) - MET793@hinge.48 (97 ) (hinge)
linker with 4 AAs PRO794@linker.49 ( 98) - SER797@linker.52 (101 ) (linker)
αD with 7 AAs LEU798@αD.53 ( 102) - GLU804@αD.59 (108 ) (αD)
αE with 5 AAs TYR827@αE.60 ( 131) - ARG831@αE.64 (135 ) (αE)
VI with 3 AAs ARG832@VI.65 ( 136) - VAL834@VI.67 (138 ) (VI)
c.l with 8 AAs HIS835@c.l.68 ( 139) - ASN842@c.l.75 (146 ) (c.l)
VII with 3 AAs VAL843@VII.76 ( 147) - VAL845@VII.78 (149 ) (VII)
VIII with 1 AAs ILE853@VIII.79 ( 157) - ILE853@VIII.79 (157 ) (VIII)
xDFG with 4 AAs THR854@xDFG.80 ( 158) - GLY857@xDFG.83 (161 ) (xDFG)
a.l with 2 AAs LEU858@a.l.84 ( 162) - ALA859@a.l.85 (163 ) (a.l)
Computing distances in the interface between fragments
0
and
1
0%| | 0/1 [00:00<?, ?it/s]
Streaming example_kinases/trajectory.7VRE.xtc (nr. 0) with stride 1 in chunks of 10000 frames. Now at chunk nr 0, frames so far 500
100%|█████████████████████████████████████████████████████████████████████| 1/1 [00:00<00:00, 1.44it/s]
These 17 contacts capture 13.02 (~97%) of the total frequency 13.39 (over 323 input contacts)
As orientation value, the first 15 ctcs already capture 90.0% of 13.39.
The 15-th contact has a frequency of 0.40
freq label residue idxs sum
0 1.00 M793@hinge.48 - 7VH1@ligand 97 323 1.00
1 1.00 L718@I.3 - 7VH1@ligand 22 323 2.00
2 0.98 V726@II.11 - 7VH1@ligand 30 323 2.97
3 0.96 P794@linker.49 - 7VH1@ligand 98 323 3.93
4 0.95 A743@III.15 - 7VH1@ligand 47 323 4.88
5 0.94 L844@VII.77 - 7VH1@ligand 148 323 5.82
6 0.92 Q791@hinge.46 - 7VH1@ligand 95 323 6.74
7 0.92 G796@linker.51 - 7VH1@ligand 100 323 7.66
8 0.91 F723@g.l.8 - 7VH1@ligand 27 323 8.57
9 0.82 L792@hinge.47 - 7VH1@ligand 96 323 9.40
10 0.72 T854@xDFG.80 - 7VH1@ligand 158 323 10.12
11 0.68 K745@III.17 - 7VH1@ligand 49 323 10.80
12 0.66 M790@GK.45 - 7VH1@ligand 94 323 11.46
13 0.54 G719@g.l.4 - 7VH1@ligand 23 323 12.00
14 0.40 D855@xDFG.81 - 7VH1@ligand 159 323 12.40
15 0.36 R841@c.l.74 - 7VH1@ligand 145 323 12.76
16 0.26 L1001@EGFR - 7VH1@ligand 305 323 13.02
label freq
0 M793@hinge.48 1.00
1 L718@I.3 1.00
2 V726@II.11 0.98
3 P794@linker.49 0.96
4 A743@III.15 0.95
5 L844@VII.77 0.94
6 Q791@hinge.46 0.92
7 G796@linker.51 0.92
8 F723@g.l.8 0.91
9 L792@hinge.47 0.82
10 T854@xDFG.80 0.72
11 K745@III.17 0.68
12 M790@GK.45 0.66
13 G719@g.l.4 0.54
14 D855@xDFG.81 0.40
15 R841@c.l.74 0.36
16 L1001@EGFR 0.26
label freq
0 7VH1@ligand 13.02
Compare interactions across the four compounds in a violinplot¶
Additionally, we will display representative geometries directly on the violinplots via their residue-residue distance-values. Subsequently, we will view these geometries in 3D
[7]:
colors = mdciao.plots.color_dict_guesser("tab10", binding_pocket.keys())
myfig, myax, keys = mdciao.plots.compare_violins(binding_pocket,
colors=colors,
anchor="ligand",
ctc_cutoff_Ang=4,
mutations_dict={
"EUX1": "ligand",
"7VH1": "ligand",
"W321": "ligand",
"P31": "ligand"
},
defrag=None,
sort_by="residue",
inch_per_contacts=.80,
legend_rows=2,
representatives=True,
)
myax.set_title("binding pocket interactions"
"\nfor 4 different EGFR inhibitors")
myfig.tight_layout()
#myfig.savefig("EGFR.png", bbox_inches="tight")
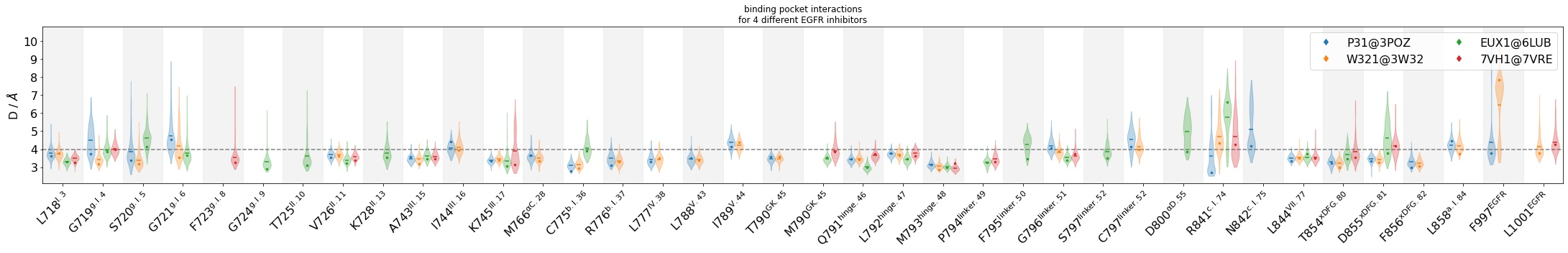
Show the representative geometries¶
These are the same geometries being shown as small dots inside the violins of the previous figure, using the repframes method:
[8]:
representatives = {}
ref = None
for key, bp in binding_pocket.items():
repframe = bp.repframes(return_traj=True)[-1][0]
representatives[key] = repframe
Returning frame 83 of traj nr. 0: example_kinases/trajectory.3POZ.xtc
Returning frame 197 of traj nr. 0: example_kinases/trajectory.3W32.xtc
Returning frame 369 of traj nr. 0: example_kinases/trajectory.6LUB.xtc
Returning frame 253 of traj nr. 0: example_kinases/trajectory.7VRE.xtc
Superpose structures using the KLIFs alignment labels¶
This way, the alignment will be particularly good in the binding pocket
[9]:
KLIFS_alignment = mdciao.nomenclature.AlignerConsensus({key : bp.top for key, bp in binding_pocket.items()},
CL=KLIFS)
KLIFS_alignment.AAresSeq
[9]:
| consensus | P31@3POZ | W321@3W32 | EUX1@6LUB | 7VH1@7VRE | |
|---|---|---|---|---|---|
| 0 | GK.45 | THR790 | THR790 | MET790 | MET790 |
| 1 | I.1 | LYS716 | LYS716 | LYS716 | LYS716 |
| 2 | I.2 | VAL717 | VAL717 | VAL717 | VAL717 |
| 3 | I.3 | LEU718 | LEU718 | LEU718 | LEU718 |
| 4 | II.10 | THR725 | THR725 | THR725 | THR725 |
| ... | ... | ... | ... | ... | ... |
| 80 | αE.60 | TYR827 | TYR827 | TYR827 | TYR827 |
| 81 | αE.61 | LEU828 | LEU828 | LEU828 | LEU828 |
| 82 | αE.62 | GLU829 | GLU829 | GLU829 | GLU829 |
| 83 | αE.63 | ASP830 | ASP830 | ASP830 | ASP830 |
| 84 | αE.64 | ARG831 | ARG831 | ARG831 | ARG831 |
85 rows × 5 columns
[10]:
# We can directly get CA indices to map atoms
KLIFS_alignment.CAidxs
[10]:
| consensus | P31@3POZ | W321@3W32 | EUX1@6LUB | 7VH1@7VRE | |
|---|---|---|---|---|---|
| 0 | GK.45 | 1435 | 1435 | 1495 | 1495 |
| 1 | I.1 | 280 | 280 | 340 | 340 |
| 2 | I.2 | 302 | 302 | 362 | 362 |
| 3 | I.3 | 318 | 318 | 378 | 378 |
| 4 | II.10 | 399 | 399 | 459 | 459 |
| ... | ... | ... | ... | ... | ... |
| 80 | αE.60 | 2029 | 2029 | 2092 | 2092 |
| 81 | αE.61 | 2050 | 2050 | 2113 | 2113 |
| 82 | αE.62 | 2069 | 2069 | 2132 | 2132 |
| 83 | αE.63 | 2084 | 2084 | 2147 | 2147 |
| 84 | αE.64 | 2096 | 2096 | 2159 | 2159 |
85 rows × 5 columns
[11]:
ref_key = "W321@3W32" # We take this one but could be any one
ref_geom = representatives[ref_key]
for key, geom in representatives.items():
if key!=ref_key:
ref_CAs, key_CAs = KLIFS_alignment.CAidxs[[ref_key, key]].values.T
geom.superpose(ref_geom, atom_indices=key_CAs, ref_atom_indices=ref_CAs)
Visualize residues with different behaviors in each compound¶
For example, residues * 775@b.l.36 * 841@c.l.74 * 855@xDFG.81 * 997@EGFR (doesn’t have a KLIFS label)
[12]:
colors = {key: matplotlib.colors.to_hex(col) for key, col in colors.items()}
iwd = nglview.NGLWidget()
for ii, (key, rep) in enumerate(representatives.items()):
iwd.add_trajectory(rep)
iwd.clear_representations(component=ii)
iwd.add_cartoon(color="white", component=ii)
iwd.add_licorice(color=colors[key], component=ii, selection="(775 841 855 997) and not Hydrogen", radius=.1)
iwd.add_ball_and_stick(color=colors[key], component=ii,
selection="not protein and not Hydrogen",
radius=.1,
)
iwd
References¶
The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 (PDB ID 7VRE)
Chen, H., Lai, M., Zhang, T., Chen, Y., Tong, L., Zhu, S., … Ding, K. (2022). Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations. Journal of Medicinal Chemistry, 65(9), 6840–6858. https://doi.org/10.1021/acs.jmedchem.2c00168
EGFR kinase domain complexed with compound 20a (PDB ID 3W32)
Kawakita, Y., Seto, M., Ohashi, T., Tamura, T., Yusa, T., Miki, H., … Ishikawa, T. (2013). Design and synthesis of novel pyrimido[4,5- b ]azepine derivatives as HER2/EGFR dual inhibitors. Bioorganic & Medicinal Chemistry, 21(8), 2250–2261. https://doi.org/10.1016/j.bmc.2013.02.014
EGFR Kinase domain complexed with tak-285 (PDB ID 3POZ)
Aertgeerts, K., Skene, R., Yano, J., Sang, B. C., Zou, H., Snell, G., … Sogabe, S. (2011). Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. Journal of Biological Chemistry, 286(21), 18756–18765. https://doi.org/10.1074/jbc.M110.206193
Crystal Structure of EGFR(L858R/T790M/C797S) in complex with CH7233163 (PDB ID 6LUB)
Kashima, K., Kawauchi, H., Tanimura, H., Tachibana, Y., Chiba, T., Torizawa, T., & Sakamoto, H. (2020). CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation. Molecular Cancer Therapeutics, 19(11), 2288–2297. https://doi.org/10.1158/1535-7163.MCT-20-0229