Missing Contacts
You might find that some contacts that you were expecting to be found by mdciao don’t actually show up in mdciao’s results. Several input parameters control the contact reporting of mdciao, and it might not be obvious which one of them (if any) is actually hiding your contact. The logic behind these parameters, and their default values, is fairly straightforward, and we illustrate it here.
If you want to run this notebook on your own, please download and extract the data from here first. You can download it:
using the browser
using the terminal with
wget http://proteinformatics.org/mdciao/mdciao_example.zip; unzip mdciao_example.zipusing mdciao’s own method mdciao.examples.fetch_example_data
If you want to take a 3D-look at this data, you can do it here.
ctc_cutoff_Ang
This is the most obvious parameter that controls the contact computation. It appears virtually in all methods (CLI or API) that compute contact frequencies. Whenever it has a default value, it is 4.5 Angstrom.
Note
Please see the note of caution on the use of hard cutoffs in the main page of the docs.
[1]:
import mdciao, os
if not os.path.exists("mdciao_example"):
mdciao.examples.fetch_example_data()
[2]:
import mdtraj as md
traj = md.load("mdciao_example/traj.xtc",top="mdciao_example/top.pdb")
First, we individually call mdciao.cli.residue_neighborhoods with two ctc_cutoff_Ang values, 3.0, 3.5, and 4.0 Angstrom. This will generate three frequency reports which we will later compare with mdciao.cli.compare. Please refer to those methods if their function calls aren’t
entirely clear to you.
Note
We are hiding the outputs with the use of the `%%capture magic <https://ipython.readthedocs.io/en/stable/interactive/magics.html#cellmagic-capture>`__.
[3]:
%%capture
for ctc_cutoff_Ang in [3, 3.5, 4.0, 4.5, 5.0]:
mdciao.cli.residue_neighborhoods("L394",traj,
short_AA_names=True,
ctc_cutoff_Ang=ctc_cutoff_Ang,
figures=False,
fragment_names=None,
#ctc_control=1.0,
no_disk=False)[353]
[4]:
mdciao.cli.compare({
"ctc_cutoff_Ang = 5.0 AA" : "neighborhood.LEU394@5.0_Ang.dat",
"ctc_cutoff_Ang = 4.5 AA" : "neighborhood.LEU394@4.5_Ang.dat",
"ctc_cutoff_Ang = 4.0 AA" : "neighborhood.LEU394@4.0_Ang.dat",
"ctc_cutoff_Ang = 3.5 AA" : "neighborhood.LEU394@3.5_Ang.dat",
"ctc_cutoff_Ang = 3.0 AA" : "neighborhood.LEU394@3.0_Ang.dat",
},
anchor="L394",
title="Cutoff comparison");
These interactions are not shared:
I233, L230, R385
Their cumulative ctc freq is 8.00.
Created files
freq_comparison.pdf
freq_comparison.xlsx
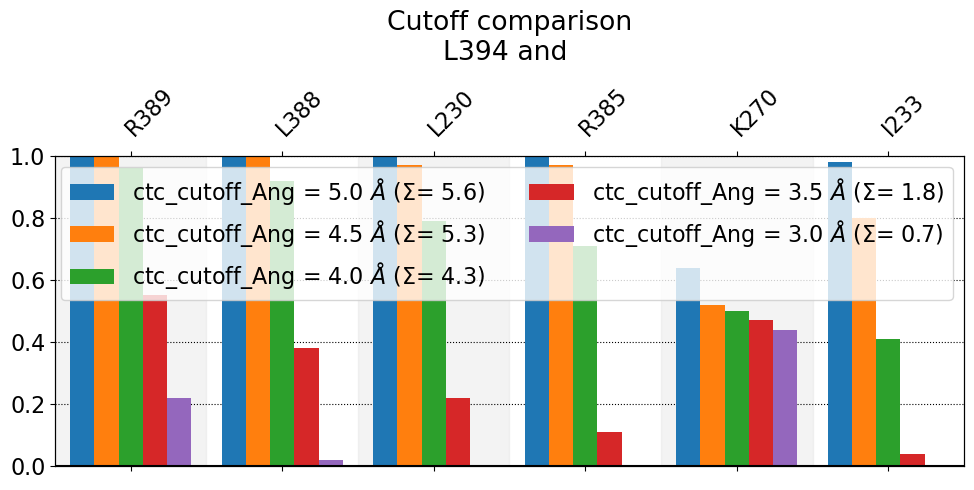
We observe that the smaller the cutoff, the fewer contacts get reported. In this case L230, R385, and I233 never approach L394 at distances smaller than 3.0 Angstrom during in the entire simulation, hence they don’t get reported (they don’t get any purple-bars). As for K270, the frequency doesn’t change very much, because it’s a salt-bridge that’s really either formed at very close distance or broken at higher distances, as can be seen in this time-trace
figure. Also notice that, the higher the cutoff, the higher the sum over bars, \(\Sigma\), since the height of the bars has increased.
ctc_control
Even when using the same ctc_cutoff_Ang, there’s other ways of controlling what gets reported. ctc_control is one of them. This parameter controls how many residues get reported per neighborhood, since usually one is not interested in all residues but only the most frequent ones.
Controlling with integers
One way to control this is to select only the first n frequent ones (n is an integer and is 6 by default). Here we do the comparison again, but withoug hiding the output s.t. you can see the contact list grow.
[5]:
#%%capture
ctc_controls = [4,5,6,7,8]
for ctc_control in ctc_controls:
mdciao.cli.residue_neighborhoods("L394",traj,
short_AA_names=True,
ctc_control=ctc_control,
figures=False,
fragment_names=None,
no_disk=False,
output_desc='neighborhood.ctc_control_%u'%ctc_control)
mdciao.cli.compare({"ctc-control = %u"%key : "neighborhood.ctc_control_%u.LEU394@4.5_Ang.dat"%(key)
for key in ctc_controls},
anchor="L394",
title="ctc-control comparison");
Will compute contact frequencies for (1 items):
<mdtraj.Trajectory with 280 frames, 8384 atoms, 1044 residues, and unitcells>
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Will compute neighborhoods for the residues
L394
excluding 4 nearest neighbors
residue residx fragment resSeq
LEU394 353 0 394
Performing a first pass on 1039 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances:
Reduced to only 43 residue pairs for the computation of actual residue-residue distances:
L394:
The following 4 contacts capture 3.94 (~72%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 6 ctcs already capture 90.0% of 5.43.
The 6-th contact has a frequency of 0.52.
freq label residues fragments sum
1 1.00 L394 - L388 353 - 347 0 - 0 1.00
2 1.00 L394 - R389 353 - 348 0 - 0 2.00
3 0.97 L394 - L230 353 - 957 0 - 3 2.97
4 0.97 L394 - R385 353 - 344 0 - 0 3.94
The following files have been created:
./neighborhood.ctc_control_4.LEU394@4.5_Ang.dat
Will compute contact frequencies for (1 items):
<mdtraj.Trajectory with 280 frames, 8384 atoms, 1044 residues, and unitcells>
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Will compute neighborhoods for the residues
L394
excluding 4 nearest neighbors
residue residx fragment resSeq
LEU394 353 0 394
Performing a first pass on 1039 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances:
Reduced to only 43 residue pairs for the computation of actual residue-residue distances:
L394:
The following 5 contacts capture 4.74 (~87%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 6 ctcs already capture 90.0% of 5.43.
The 6-th contact has a frequency of 0.52.
freq label residues fragments sum
1 1.00 L394 - L388 353 - 347 0 - 0 1.00
2 1.00 L394 - R389 353 - 348 0 - 0 2.00
3 0.97 L394 - L230 353 - 957 0 - 3 2.97
4 0.97 L394 - R385 353 - 344 0 - 0 3.94
5 0.80 L394 - I233 353 - 960 0 - 3 4.74
The following files have been created:
./neighborhood.ctc_control_5.LEU394@4.5_Ang.dat
Will compute contact frequencies for (1 items):
<mdtraj.Trajectory with 280 frames, 8384 atoms, 1044 residues, and unitcells>
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Will compute neighborhoods for the residues
L394
excluding 4 nearest neighbors
residue residx fragment resSeq
LEU394 353 0 394
Performing a first pass on 1039 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances:
Reduced to only 43 residue pairs for the computation of actual residue-residue distances:
L394:
The following 6 contacts capture 5.26 (~97%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 6 ctcs already capture 90.0% of 5.43.
The 6-th contact has a frequency of 0.52.
freq label residues fragments sum
1 1.00 L394 - L388 353 - 347 0 - 0 1.00
2 1.00 L394 - R389 353 - 348 0 - 0 2.00
3 0.97 L394 - L230 353 - 957 0 - 3 2.97
4 0.97 L394 - R385 353 - 344 0 - 0 3.94
5 0.80 L394 - I233 353 - 960 0 - 3 4.74
6 0.52 L394 - K270 353 - 972 0 - 3 5.26
The following files have been created:
./neighborhood.ctc_control_6.LEU394@4.5_Ang.dat
Will compute contact frequencies for (1 items):
<mdtraj.Trajectory with 280 frames, 8384 atoms, 1044 residues, and unitcells>
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Will compute neighborhoods for the residues
L394
excluding 4 nearest neighbors
residue residx fragment resSeq
LEU394 353 0 394
Performing a first pass on 1039 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances:
Reduced to only 43 residue pairs for the computation of actual residue-residue distances:
L394:
The following 7 contacts capture 5.39 (~99%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 6 ctcs already capture 90.0% of 5.43.
The 6-th contact has a frequency of 0.52.
freq label residues fragments sum
1 1.00 L394 - L388 353 - 347 0 - 0 1.00
2 1.00 L394 - R389 353 - 348 0 - 0 2.00
3 0.97 L394 - L230 353 - 957 0 - 3 2.97
4 0.97 L394 - R385 353 - 344 0 - 0 3.94
5 0.80 L394 - I233 353 - 960 0 - 3 4.74
6 0.52 L394 - K270 353 - 972 0 - 3 5.26
7 0.13 L394 - K267 353 - 969 0 - 3 5.39
The following files have been created:
./neighborhood.ctc_control_7.LEU394@4.5_Ang.dat
Will compute contact frequencies for (1 items):
<mdtraj.Trajectory with 280 frames, 8384 atoms, 1044 residues, and unitcells>
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Will compute neighborhoods for the residues
L394
excluding 4 nearest neighbors
residue residx fragment resSeq
LEU394 353 0 394
Performing a first pass on 1039 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances:
Reduced to only 43 residue pairs for the computation of actual residue-residue distances:
L394:
The following 8 contacts capture 5.41 (~100%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 6 ctcs already capture 90.0% of 5.43.
The 6-th contact has a frequency of 0.52.
freq label residues fragments sum
1 1.00 L394 - L388 353 - 347 0 - 0 1.00
2 1.00 L394 - R389 353 - 348 0 - 0 2.00
3 0.97 L394 - L230 353 - 957 0 - 3 2.97
4 0.97 L394 - R385 353 - 344 0 - 0 3.94
5 0.80 L394 - I233 353 - 960 0 - 3 4.74
6 0.52 L394 - K270 353 - 972 0 - 3 5.26
7 0.13 L394 - K267 353 - 969 0 - 3 5.39
8 0.02 L394 - Q229 353 - 956 0 - 3 5.41
The following files have been created:
./neighborhood.ctc_control_8.LEU394@4.5_Ang.dat
These interactions are not shared:
I233, K267, K270, Q229
Their cumulative ctc freq is 5.04.
Created files
freq_comparison.pdf
freq_comparison.xlsx
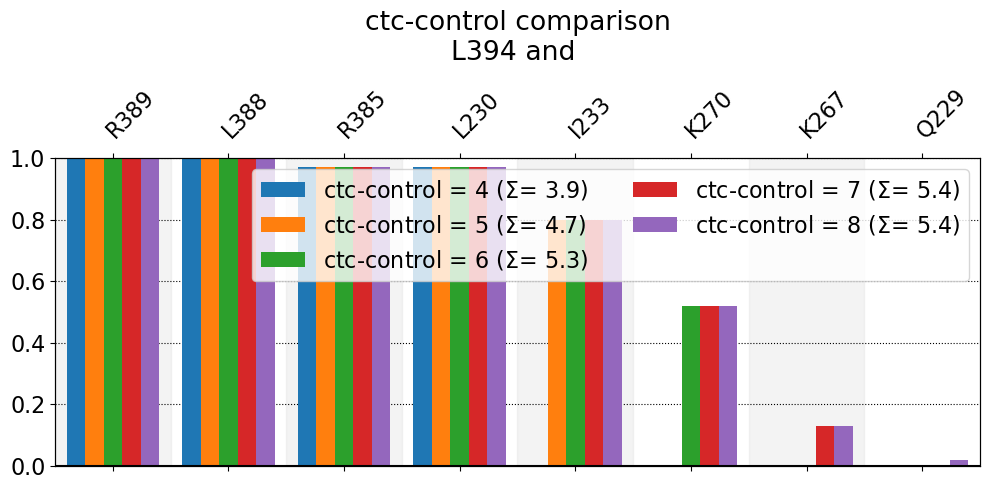
n is simply the number of reported bars: four blue ones, five orange ones, six green ones, seven red ones and eight purple ones. \(\Sigma\) is just the sum of the heights of all bars and is thus an estimate of the average number of neighbors that are being reported (at this cutoff). A couple of observations:
The relation of \(\Sigma\) with n is straightforward: as n grows, so does \(\Sigma\), although it grows slower with each n until it barely changes, given that the last bars it sums over are negligible. In other words, summing over the last purple bar (barely visible) for
Q229doesn’t really change \(\Sigma\) much.Hence, for a fixed cutoff, there’s an upper bond to \(\Sigma\) (\(\Sigma\leq\Sigma_t\)), because the total average number of neighbors, \(\Sigma_t\), of a given residue, over a given dataset, is determined by the used cutoff only. The
ctc_controlparameter simply determines how many neighbors get actually reported.Please note that for very small datasets, like one single pdb file, the word frequency doesn’t really have a defined meaning, so that, for this cases, it is better to use
ctc_control=1.0(more on this below).In cases where
mdciaois used to look for the neighborhood of just one residue, there’s a sensible number of residues to choose (somewhere between 5 and 10), because usually that’s how many interactions a residue will have (please note, this doesn’t necessary hold for elongated ligands, lipids, acid chains etc).
mdciao writes out the percentage of \(\Sigma_t\) captured by the reported contacts, so that the user can decide whether to increase n or not. For each of the iterations above, here are terminal-output reporting precisely that:
ctc_control = 4The following 4 contacts capture 3.94 (~72%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom). As orientation value, the first 6 ctcs already capture 90.0% of 5.43. The 6-th contact has a frequency of 0.52. freq label residues fragments sum 1 1.00 L394 - L388 353 - 347 0 - 0 1.00 2 1.00 L394 - R389 353 - 348 0 - 0 2.00 3 0.97 L394 - L230 353 - 957 0 - 3 2.97 4 0.97 L394 - R385 353 - 344 0 - 0 3.94The intention is to report how much of \(\Sigma_t\) has been captured using 3 contacts (~72%), and how many would be needed to capture most (90%) of it (6 contacts). The actual value \(\Sigma_t\) is reported immediately after (total frequency 5.43). You will notice that the value 5.43 doesn’t change as we increase n in the reports below. The 9 contacts with nonzero frequency at 4.50 Angstrom is just telling that only 9 residue-pairs were closer than distance the distance cutoff in at least %1 of the data (see the parametermin_freq=0.01).
So, as we increase n, the listed contacts grows and the reported percentage grows, but all the rest stays the same
ctc_control = 5The following 5 contacts capture 4.74 (~87%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom). As orientation value, the first 6 ctcs already capture 90.0% of 5.43. The 6-th contact has a frequency of 0.52. freq label residues fragments sum 1 1.00 L394 - L388 353 - 347 0 - 0 1.00 2 1.00 L394 - R389 353 - 348 0 - 0 2.00 3 0.97 L394 - L230 353 - 957 0 - 3 2.97 4 0.97 L394 - R385 353 - 344 0 - 0 3.94 5 0.80 L394 - I233 353 - 960 0 - 3 4.74ctc_control = 6The following 6 contacts capture 5.26 (~97%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom). As orientation value, the first 6 ctcs already capture 90.0% of 5.43. The 6-th contact has a frequency of 0.52. freq label residues fragments sum 1 1.00 L394 - L388 353 - 347 0 - 0 1.00 2 1.00 L394 - R389 353 - 348 0 - 0 2.00 3 0.97 L394 - L230 353 - 957 0 - 3 2.97 4 0.97 L394 - R385 353 - 344 0 - 0 3.94 5 0.80 L394 - I233 353 - 960 0 - 3 4.74 6 0.52 L394 - K270 353 - 972 0 - 3 5.26ctc_control = 7The following 7 contacts capture 5.39 (~99%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom). As orientation value, the first 6 ctcs already capture 90.0% of 5.43. The 6-th contact has a frequency of 0.52. freq label residues fragments sum 1 1.00 L394 - L388 353 - 347 0 - 0 1.00 2 1.00 L394 - R389 353 - 348 0 - 0 2.00 3 0.97 L394 - L230 353 - 957 0 - 3 2.97 4 0.97 L394 - R385 353 - 344 0 - 0 3.94 5 0.80 L394 - I233 353 - 960 0 - 3 4.74 6 0.52 L394 - K270 353 - 972 0 - 3 5.26 7 0.13 L394 - K267 353 - 969 0 - 3 5.39ctc_control = 8The following 8 contacts capture 5.41 (~100%) of the total frequency 5.43 (over 9 contacts with nonzero frequency at 4.50 Angstrom). As orientation value, the first 6 ctcs already capture 90.0% of 5.43. The 6-th contact has a frequency of 0.52. freq label residues fragments sum 1 1.00 L394 - L388 353 - 347 0 - 0 1.00 2 1.00 L394 - R389 353 - 348 0 - 0 2.00 3 0.97 L394 - L230 353 - 957 0 - 3 2.97 4 0.97 L394 - R385 353 - 344 0 - 0 3.94 5 0.80 L394 - I233 353 - 960 0 - 3 4.74 6 0.52 L394 - K270 353 - 972 0 - 3 5.26 7 0.13 L394 - K267 353 - 969 0 - 3 5.39 8 0.02 L394 - Q229 353 - 956 0 - 3 5.41
Controlling with fractions
Additionally, the upper bond itself, \(\Sigma_t\), can be used as a reference for controlling how many contacts get reported. So, instead of specifying an absolute number (n = 4,5,6…), one can specify a relative number, i.e. a fraction of \(\Sigma_t\) to be reported.
This way, regardless of the actual number of involved contacts, one can choose to report most of them. In the example below, we choose to report 90% of the total number of average neighbors, which, as we know from above, is achieved with 6 contacts. To avoid introducing a new parameter, mdciao knows that if ctc_control is an integer (1, 2, 3…), the user means these many contacts, whereas if ctc_control is between 0 and 1 (.50 for 50%, .9 for 90% etc) the user wants that
fraction of \(\Sigma_t\):
[6]:
mdciao.cli.residue_neighborhoods("L394",traj,
short_AA_names=True,
ctc_cutoff_Ang=ctc_cutoff_Ang,
ctc_control=1.0,
figures=False,
fragment_names=None,
no_disk=False,
);
Will compute contact frequencies for (1 items):
<mdtraj.Trajectory with 280 frames, 8384 atoms, 1044 residues, and unitcells>
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Will compute neighborhoods for the residues
L394
excluding 4 nearest neighbors
residue residx fragment resSeq
LEU394 353 0 394
Performing a first pass on 1039 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances:
Reduced to only 45 residue pairs for the computation of actual residue-residue distances:
L394:
The following 9 contacts capture 6.09 (~100%) of the total frequency 6.09 (over 11 contacts with nonzero frequency at 5.00 Angstrom).
As orientation value, the first 6 ctcs already capture 90.0% of 6.09.
The 6-th contact has a frequency of 0.64.
freq label residues fragments sum
1 1.00 L394 - R389 353 - 348 0 - 0 1.00
2 1.00 L394 - L388 353 - 347 0 - 0 2.00
3 1.00 L394 - L230 353 - 957 0 - 3 3.00
4 1.00 L394 - R385 353 - 344 0 - 0 3.99
5 0.98 L394 - I233 353 - 960 0 - 3 4.97
6 0.64 L394 - K270 353 - 972 0 - 3 5.60
7 0.36 L394 - K267 353 - 969 0 - 3 5.97
8 0.09 L394 - Q229 353 - 956 0 - 3 6.05
9 0.03 L394 - A226 353 - 953 0 - 3 6.09
The following files have been created:
./neighborhood.LEU394@5.0_Ang.dat
Which control-mode to choose (fraction or integers) is up to the user:
For compactness, limiting the output with integers can avoid huge outputs, but might leave out some interesting stuff.
For completeness, choosing
ctc_control = 1.0will ensure that any-residue under the cutoff (regardless of its frequency) will be reported, but might fill the reports with “long tails” of many residues with very low frequencies (see below).
n_nearest
Another way in which mdciao is choosing which contacts to show is through excluding nearest neighbors, i.e. residues that are predictably going to be under the cutoff simply because they are bonded neighbors of the residue in question.
In our example, for L394 these would be residues 395 and 393 (first bonded) 396 and 392 (second bonded) and so on (except, in this particular L394 case, since it’s a terminal residue, we only get 393, 392 etc).
The default value that mdciao has for this parameter is n_nearest=4, which is approximately one \(\alpha\)-helix turn away. The intention is to avoid reporting residues in the helix-turns immediately before and immediately after the residue.
Let’s see the effect of varying this parameter:
[7]:
%%capture
n_nearests = [0, 1, 2, 3, 4]
for n_nearest in n_nearests:
n = mdciao.cli.residue_neighborhoods("L394",traj,
short_AA_names=True,
ctc_control=20,
n_nearest=n_nearest,
figures=False,
no_disk=False,
output_desc='neighborhood.n_nearest_%u'%n_nearest)
[8]:
mdciao.cli.compare({"n_n = %u"%key : "neighborhood.n_nearest_%u.LEU394@frag0@%2.1f_Ang.dat"%(key,4.5)
for key in n_nearests},
anchor="L394",
sort_by="residue",
title="n_neighbors comparison");
These interactions are not shared:
E392, L393
Their cumulative ctc freq is 3.00.
Created files
freq_comparison.pdf
freq_comparison.xlsx
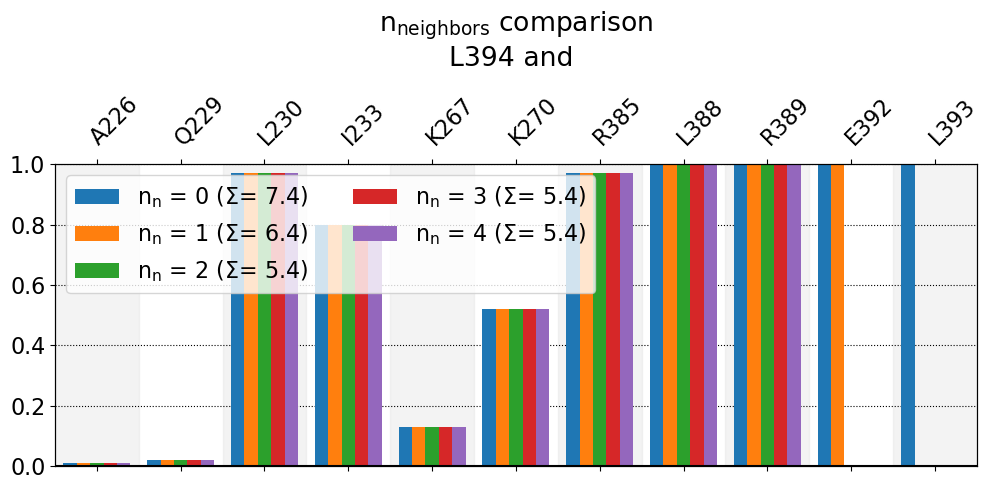
- The bars:Since we’ve sorted the frequency bars by increasing residue number, the closer to the right the bar is, the closer (in space) to
L394we are. Hence,L394’s immediate bonded neighbor,L393, only gets a bar whenn=0(no excuded neighbors, blue bar). Accordingly, next the residue after that,E392, only gets a bar withn=0orn=1, else it’s excluded. SinceL393is covalently bonded toL394, andL392is covalently bonded toL392, there’s a strong expectation for these residue to be near each other, so these frequencies are not very informative. If you’re wondering what’s with positions 390 and 391, there’s a bend in the C-terminus of the alpha helix 5 (TYR391points away fromL394throughout the simulation) so they do not appear on the report regardless. - The legend:We can see also here that, the lower
n_neighbors, i.e. the less neighbors we exclude, the higher the \(\Sigma\) value. So, asn_neighborsgoes up, these two bars get hidden, and the graph doesn’t change anymore.
Interfaces: ctc_control and min_freq
When computing interfaces between two different groups of residues using mdciao.cli.interface, one can set ctc_control=1.0 and min_freq=O to force mdciao to report all nonzero frequencies. This means any and all residue pairs that, at any given point in time, might have been at a distance >= ctc_cutoff_Ang (even if was just for one frame) will be
reported.
After reporting the most relevant contacts, this approach typically reports also a high number of very low frequency contacts, i.e. a long tail of very low frequency contacts, in particular for large interfaces (more on this tail below).
First, we take a look at the full list of contacts by using: * ctc_control=1.0 * min_freq=0
[9]:
intf = mdciao.cli.interface("mdciao_example/traj.xtc", topology="mdciao_example/top.pdb",
no_disk=True, interface_selection_1=[0], interface_selection_2=[1],
ctc_control=1.0,
min_freq=0,
figures=False);
intf.plot_freqs_as_bars(4.5, shorten_AAs=True, defrag="@", cumsum=True);
Will compute contact frequencies for trajectories:
mdciao_example/traj.xtc
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Select group 1: 0
Select group 2: 1
Will look for contacts in the interface between fragments
0
and
1.
Performing a first pass on the 120360 group_1-group_2 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances.
Reduced to only 1153 (from 120360) residue pairs for the computation of actual residue-residue distances:
The following 138 contacts capture 74.50 (~100%) of the total frequency 74.50 (over 138 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 73 ctcs already capture 90.0% of 74.50.
The 73-th contact has a frequency of 0.57.
freq label residues fragments sum
1 1.00 I26 - K89 22 - 442 0 - 1 1.00
2 1.00 R228 - G162 194 - 515 0 - 1 2.00
3 1.00 R228 - D186 194 - 539 0 - 1 3.00
4 1.00 Q227 - Y145 193 - 498 0 - 1 4.00
5 1.00 Y37 - L55 33 - 408 0 - 1 5.00
6 1.00 K233 - N230 199 - 583 0 - 1 6.00
7 1.00 K233 - D246 199 - 599 0 - 1 7.00
8 1.00 D33 - K78 29 - 431 0 - 1 8.00
9 1.00 F238 - L117 204 - 470 0 - 1 9.00
10 1.00 F238 - W99 204 - 452 0 - 1 10.00
11 1.00 Q227 - N119 193 - 472 0 - 1 11.00
12 1.00 L30 - G53 26 - 406 0 - 1 12.00
13 1.00 R201 - R96 169 - 449 0 - 1 13.00
14 1.00 N23 - N88 19 - 441 0 - 1 14.00
15 1.00 N23 - K89 19 - 442 0 - 1 15.00
16 1.00 F222 - L117 188 - 470 0 - 1 16.00
17 1.00 F222 - W99 188 - 452 0 - 1 17.00
18 1.00 C237 - W99 203 - 452 0 - 1 18.00
19 1.00 E27 - K89 23 - 442 0 - 1 19.00
20 1.00 C237 - Q75 203 - 428 0 - 1 20.00
21 1.00 C237 - K57 203 - 410 0 - 1 21.00
22 1.00 I26 - A92 22 - 445 0 - 1 22.00
23 1.00 D240 - K57 206 - 410 0 - 1 23.00
24 1.00 Q236 - Y59 202 - 412 0 - 1 24.00
25 1.00 K233 - D290 199 - 643 0 - 1 24.99
26 1.00 C237 - Y59 203 - 412 0 - 1 25.99
27 1.00 Q236 - W332 202 - 685 0 - 1 26.99
28 1.00 Q19 - N88 15 - 441 0 - 1 27.98
29 0.99 I172 - E130 140 - 483 0 - 1 28.97
30 0.99 Q227 - L117 193 - 470 0 - 1 29.97
31 0.99 R20 - N88 16 - 441 0 - 1 30.96
32 0.99 K34 - L55 30 - 408 0 - 1 31.95
33 0.99 Q236 - K57 202 - 410 0 - 1 32.94
34 0.99 Q19 - T86 15 - 439 0 - 1 33.92
35 0.98 A22 - K89 18 - 442 0 - 1 34.90
36 0.98 R228 - Y145 194 - 498 0 - 1 35.89
37 0.98 F208 - D118 174 - 471 0 - 1 36.87
38 0.98 C237 - L117 203 - 470 0 - 1 37.85
39 0.98 W281 - R314 240 - 667 0 - 1 38.82
40 0.98 I172 - N132 140 - 485 0 - 1 39.80
41 0.98 W234 - Y145 200 - 498 0 - 1 40.77
42 0.97 Q19 - D83 15 - 436 0 - 1 41.74
43 0.97 R42 - W99 38 - 452 0 - 1 42.71
44 0.97 R42 - D76 38 - 429 0 - 1 43.69
45 0.96 D33 - L55 29 - 408 0 - 1 44.65
46 0.96 W234 - L117 200 - 470 0 - 1 45.61
47 0.96 I26 - H91 22 - 444 0 - 1 46.58
48 0.96 E209 - S98 175 - 451 0 - 1 47.54
49 0.94 I172 - R134 140 - 487 0 - 1 48.47
50 0.93 F208 - L117 174 - 470 0 - 1 49.41
51 0.93 Q227 - G144 193 - 497 0 - 1 50.34
52 0.93 N239 - W332 205 - 685 0 - 1 51.27
53 0.92 C237 - M101 203 - 454 0 - 1 52.19
54 0.91 Q19 - R68 15 - 421 0 - 1 53.10
55 0.90 Q19 - V90 15 - 443 0 - 1 54.01
56 0.90 Q170 - R129 138 - 482 0 - 1 54.91
57 0.89 G226 - T143 192 - 496 0 - 1 55.80
58 0.89 W281 - W332 240 - 685 0 - 1 56.69
59 0.87 F208 - N119 174 - 472 0 - 1 57.56
60 0.86 L30 - K89 26 - 442 0 - 1 58.42
61 0.83 Q170 - G131 138 - 484 0 - 1 59.25
62 0.81 A22 - V90 18 - 443 0 - 1 60.06
63 0.80 I26 - V90 22 - 443 0 - 1 60.86
64 0.77 I172 - G131 140 - 484 0 - 1 61.63
65 0.75 E16 - T86 12 - 439 0 - 1 62.39
66 0.70 K233 - Y59 199 - 412 0 - 1 63.09
67 0.66 Q170 - E130 138 - 483 0 - 1 63.75
68 0.63 F222 - D118 188 - 471 0 - 1 64.38
69 0.63 I207 - D118 173 - 471 0 - 1 65.01
70 0.62 L30 - L55 26 - 408 0 - 1 65.63
71 0.61 E15 - R68 11 - 421 0 - 1 66.24
72 0.59 E16 - N88 12 - 441 0 - 1 66.83
73 0.57 W234 - M101 200 - 454 0 - 1 67.40
74 0.54 R232 - D228 198 - 581 0 - 1 67.94
75 0.52 E209 - W99 175 - 452 0 - 1 68.46
76 0.50 Q236 - D290 202 - 643 0 - 1 68.96
77 0.41 E209 - R96 175 - 449 0 - 1 69.38
78 0.39 Q19 - K89 15 - 442 0 - 1 69.76
79 0.37 V241 - W99 207 - 452 0 - 1 70.13
80 0.30 H220 - W99 186 - 452 0 - 1 70.44
81 0.29 L171 - E130 139 - 483 0 - 1 70.72
82 0.26 I207 - I120 173 - 473 0 - 1 70.99
83 0.23 R20 - T86 16 - 439 0 - 1 71.22
84 0.22 K233 - G272 199 - 625 0 - 1 71.44
85 0.22 R201 - E138 169 - 491 0 - 1 71.65
86 0.21 Q227 - T143 193 - 496 0 - 1 71.86
87 0.19 G226 - N119 192 - 472 0 - 1 72.05
88 0.17 R42 - A56 38 - 409 0 - 1 72.22
89 0.16 Q12 - T86 8 - 439 0 - 1 72.37
90 0.14 R42 - L55 38 - 408 0 - 1 72.51
91 0.14 L43 - W99 39 - 452 0 - 1 72.65
92 0.13 R232 - D246 198 - 599 0 - 1 72.78
93 0.12 R232 - C204 198 - 557 0 - 1 72.90
94 0.10 E101 - N132 69 - 485 0 - 1 73.00
95 0.10 E15 - T86 11 - 439 0 - 1 73.10
96 0.09 R42 - S98 38 - 451 0 - 1 73.19
97 0.09 Q12 - Y85 8 - 438 0 - 1 73.28
98 0.09 D173 - R134 141 - 487 0 - 1 73.36
99 0.08 R42 - Q75 38 - 428 0 - 1 73.44
100 0.06 Q170 - T128 138 - 481 0 - 1 73.51
101 0.06 Q12 - R68 8 - 421 0 - 1 73.57
102 0.06 N239 - K57 205 - 410 0 - 1 73.63
103 0.06 N239 - N313 205 - 666 0 - 1 73.69
104 0.05 R228 - D163 194 - 516 0 - 1 73.74
105 0.05 L30 - R52 26 - 405 0 - 1 73.79
106 0.05 V224 - L117 190 - 470 0 - 1 73.84
107 0.05 Q19 - L69 15 - 422 0 - 1 73.88
108 0.04 R20 - T87 16 - 440 0 - 1 73.92
109 0.04 W234 - Y59 200 - 412 0 - 1 73.97
110 0.04 K233 - I273 199 - 626 0 - 1 74.01
111 0.04 G226 - G144 192 - 497 0 - 1 74.05
112 0.04 R232 - D186 198 - 539 0 - 1 74.10
113 0.04 C174 - R134 142 - 487 0 - 1 74.14
114 0.04 C174 - N132 142 - 485 0 - 1 74.18
115 0.04 F238 - K57 204 - 410 0 - 1 74.21
116 0.04 I26 - I80 22 - 433 0 - 1 74.25
117 0.03 K233 - M188 199 - 541 0 - 1 74.28
118 0.03 D173 - E130 141 - 483 0 - 1 74.31
119 0.03 F222 - S98 188 - 451 0 - 1 74.34
120 0.01 R201 - L95 169 - 448 0 - 1 74.35
121 0.01 K233 - M101 199 - 454 0 - 1 74.37
122 0.01 Q236 - R314 202 - 667 0 - 1 74.38
123 0.01 N23 - T87 19 - 440 0 - 1 74.40
124 0.01 Q170 - N132 138 - 485 0 - 1 74.41
125 0.01 N167 - R129 135 - 482 0 - 1 74.43
126 0.01 L171 - R129 139 - 482 0 - 1 74.44
127 0.01 R232 - Y145 198 - 498 0 - 1 74.45
128 0.01 M221 - W99 187 - 452 0 - 1 74.45
129 0.01 K34 - G53 30 - 406 0 - 1 74.46
130 0.01 E209 - S97 175 - 450 0 - 1 74.47
131 0.01 R280 - R314 239 - 667 0 - 1 74.48
132 0.01 D240 - A56 206 - 409 0 - 1 74.48
133 0.00 R228 - G144 194 - 497 0 - 1 74.49
134 0.00 Q29 - A92 25 - 445 0 - 1 74.49
135 0.00 L30 - I80 26 - 433 0 - 1 74.49
136 0.00 D240 - W99 206 - 452 0 - 1 74.50
137 0.00 Q236 - Q75 202 - 428 0 - 1 74.50
138 0.00 L171 - R134 139 - 487 0 - 1 74.50
label freq
1 C237 5.90
2 Q19 5.20
3 Q227 4.13
4 K233 4.01
5 I26 3.80
6 I172 3.68
7 Q236 3.50
8 R228 3.04
9 F208 2.79
10 F222 2.66
11 W234 2.55
12 L30 2.54
13 Q170 2.46
14 R42 2.42
15 F238 2.04
16 N23 2.01
17 D33 1.96
18 E209 1.90
19 W281 1.86
20 A22 1.79
21 E16 1.34
22 R20 1.26
23 R201 1.23
24 G226 1.12
25 N239 1.04
26 D240 1.01
27 Y37 1.00
28 E27 1.00
29 K34 1.00
30 I207 0.90
31 R232 0.84
32 E15 0.71
33 V241 0.37
34 Q12 0.31
35 L171 0.30
36 H220 0.30
37 L43 0.14
38 D173 0.11
39 E101 0.10
40 C174 0.08
41 V224 0.05
42 N167 0.01
43 M221 0.01
44 R280 0.01
45 Q29 0.00
label freq
1 L117 5.91
2 W99 5.31
3 K89 5.23
4 L55 3.71
5 N88 3.58
6 K57 3.08
7 Y145 2.97
8 W332 2.81
9 Y59 2.74
10 V90 2.51
11 D118 2.25
12 T86 2.22
13 N119 2.06
14 E130 1.97
15 G131 1.60
16 R68 1.59
17 M101 1.51
18 D290 1.50
19 R96 1.41
20 D246 1.13
21 N132 1.13
22 T143 1.10
23 Q75 1.08
24 S98 1.08
25 R134 1.07
26 D186 1.04
27 G53 1.01
28 A92 1.00
29 G162 1.00
30 N230 1.00
31 K78 1.00
32 R314 1.00
33 G144 0.98
34 D83 0.97
35 D76 0.97
36 H91 0.96
37 R129 0.92
38 D228 0.54
39 I120 0.26
40 G272 0.22
41 E138 0.22
42 A56 0.18
43 C204 0.12
44 Y85 0.09
45 T128 0.06
46 N313 0.06
47 T87 0.06
48 D163 0.05
49 R52 0.05
50 L69 0.05
51 I273 0.04
52 I80 0.04
53 M188 0.03
54 L95 0.01
55 S97 0.01

mdciao tries to provide hints as to what might be sane values, e.g, the log reports:
The following 138 contacts capture 74.50 (~100%) of the total frequency 74.50 (over 138 contacts with nonzero frequency at 4.50 Angstrom). As orientation value, the first 73 ctcs already capture 90.0% of 74.50. The 73-th contact has a frequency of 0.57 which means: 1. the reported contacts, which have resulted from using ctc_control=1.0, min_freq=0.0, are capturing 100% of anything there is to report.mdciao looks for how many contacts one would need to capture 90% of that 100%, and the in this case the number is 73.mdciao informs the user of the frequency value of that last contact at 90%. In this case, that value (0.57 i.e. 57%) is, in this case, too high to be discarded.Hence, as a compromise between reporting everything or risking truncating too early, mdciao sets by default min_freq=0.10, i.e. contacts formed less than 10% of the time are simply not included in the returned ContactGroup. The long tail is contained in the above report, nevertheless:
[10]:
intf = mdciao.cli.interface("mdciao_example/traj.xtc", topology="mdciao_example/top.pdb",
no_disk=True, interface_selection_1=[0], interface_selection_2=[1],
ctc_control=1.0,
figures=False);
intf.plot_freqs_as_bars(4.5, shorten_AAs=True, defrag="@", cumsum=True);
Will compute contact frequencies for trajectories:
mdciao_example/traj.xtc
with a stride of 1 frames
Using method 'lig_resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 283 AAs GLU30 ( 760) - LEU340 (1042) (3) resSeq jumps
fragment 4 with 1 AAs P0G395 (1043) - P0G395 (1043) (4)
Select group 1: 0
Select group 2: 1
Will look for contacts in the interface between fragments
0
and
1.
Performing a first pass on the 120360 group_1-group_2 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances.
Reduced to only 1153 (from 120360) residue pairs for the computation of actual residue-residue distances:
The following 93 contacts capture 72.90 (~98%) of the total frequency 74.50 (over 138 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 73 ctcs already capture 90.0% of 74.50.
The 73-th contact has a frequency of 0.57.
freq label residues fragments sum
1 1.00 I26 - K89 22 - 442 0 - 1 1.00
2 1.00 R228 - G162 194 - 515 0 - 1 2.00
3 1.00 R228 - D186 194 - 539 0 - 1 3.00
4 1.00 Q227 - Y145 193 - 498 0 - 1 4.00
5 1.00 Y37 - L55 33 - 408 0 - 1 5.00
6 1.00 K233 - N230 199 - 583 0 - 1 6.00
7 1.00 K233 - D246 199 - 599 0 - 1 7.00
8 1.00 D33 - K78 29 - 431 0 - 1 8.00
9 1.00 F238 - L117 204 - 470 0 - 1 9.00
10 1.00 F238 - W99 204 - 452 0 - 1 10.00
11 1.00 Q227 - N119 193 - 472 0 - 1 11.00
12 1.00 L30 - G53 26 - 406 0 - 1 12.00
13 1.00 R201 - R96 169 - 449 0 - 1 13.00
14 1.00 N23 - N88 19 - 441 0 - 1 14.00
15 1.00 N23 - K89 19 - 442 0 - 1 15.00
16 1.00 F222 - L117 188 - 470 0 - 1 16.00
17 1.00 F222 - W99 188 - 452 0 - 1 17.00
18 1.00 C237 - W99 203 - 452 0 - 1 18.00
19 1.00 E27 - K89 23 - 442 0 - 1 19.00
20 1.00 C237 - Q75 203 - 428 0 - 1 20.00
21 1.00 C237 - K57 203 - 410 0 - 1 21.00
22 1.00 I26 - A92 22 - 445 0 - 1 22.00
23 1.00 D240 - K57 206 - 410 0 - 1 23.00
24 1.00 Q236 - Y59 202 - 412 0 - 1 24.00
25 1.00 K233 - D290 199 - 643 0 - 1 24.99
26 1.00 C237 - Y59 203 - 412 0 - 1 25.99
27 1.00 Q236 - W332 202 - 685 0 - 1 26.99
28 1.00 Q19 - N88 15 - 441 0 - 1 27.98
29 0.99 I172 - E130 140 - 483 0 - 1 28.97
30 0.99 Q227 - L117 193 - 470 0 - 1 29.97
31 0.99 R20 - N88 16 - 441 0 - 1 30.96
32 0.99 K34 - L55 30 - 408 0 - 1 31.95
33 0.99 Q236 - K57 202 - 410 0 - 1 32.94
34 0.99 Q19 - T86 15 - 439 0 - 1 33.92
35 0.98 A22 - K89 18 - 442 0 - 1 34.90
36 0.98 R228 - Y145 194 - 498 0 - 1 35.89
37 0.98 F208 - D118 174 - 471 0 - 1 36.87
38 0.98 C237 - L117 203 - 470 0 - 1 37.85
39 0.98 W281 - R314 240 - 667 0 - 1 38.82
40 0.98 I172 - N132 140 - 485 0 - 1 39.80
41 0.98 W234 - Y145 200 - 498 0 - 1 40.77
42 0.97 Q19 - D83 15 - 436 0 - 1 41.74
43 0.97 R42 - W99 38 - 452 0 - 1 42.71
44 0.97 R42 - D76 38 - 429 0 - 1 43.69
45 0.96 D33 - L55 29 - 408 0 - 1 44.65
46 0.96 W234 - L117 200 - 470 0 - 1 45.61
47 0.96 I26 - H91 22 - 444 0 - 1 46.58
48 0.96 E209 - S98 175 - 451 0 - 1 47.54
49 0.94 I172 - R134 140 - 487 0 - 1 48.47
50 0.93 F208 - L117 174 - 470 0 - 1 49.41
51 0.93 Q227 - G144 193 - 497 0 - 1 50.34
52 0.93 N239 - W332 205 - 685 0 - 1 51.27
53 0.92 C237 - M101 203 - 454 0 - 1 52.19
54 0.91 Q19 - R68 15 - 421 0 - 1 53.10
55 0.90 Q19 - V90 15 - 443 0 - 1 54.01
56 0.90 Q170 - R129 138 - 482 0 - 1 54.91
57 0.89 G226 - T143 192 - 496 0 - 1 55.80
58 0.89 W281 - W332 240 - 685 0 - 1 56.69
59 0.87 F208 - N119 174 - 472 0 - 1 57.56
60 0.86 L30 - K89 26 - 442 0 - 1 58.42
61 0.83 Q170 - G131 138 - 484 0 - 1 59.25
62 0.81 A22 - V90 18 - 443 0 - 1 60.06
63 0.80 I26 - V90 22 - 443 0 - 1 60.86
64 0.77 I172 - G131 140 - 484 0 - 1 61.63
65 0.75 E16 - T86 12 - 439 0 - 1 62.39
66 0.70 K233 - Y59 199 - 412 0 - 1 63.09
67 0.66 Q170 - E130 138 - 483 0 - 1 63.75
68 0.63 F222 - D118 188 - 471 0 - 1 64.38
69 0.63 I207 - D118 173 - 471 0 - 1 65.01
70 0.62 L30 - L55 26 - 408 0 - 1 65.63
71 0.61 E15 - R68 11 - 421 0 - 1 66.24
72 0.59 E16 - N88 12 - 441 0 - 1 66.83
73 0.57 W234 - M101 200 - 454 0 - 1 67.40
74 0.54 R232 - D228 198 - 581 0 - 1 67.94
75 0.52 E209 - W99 175 - 452 0 - 1 68.46
76 0.50 Q236 - D290 202 - 643 0 - 1 68.96
77 0.41 E209 - R96 175 - 449 0 - 1 69.38
78 0.39 Q19 - K89 15 - 442 0 - 1 69.76
79 0.37 V241 - W99 207 - 452 0 - 1 70.13
80 0.30 H220 - W99 186 - 452 0 - 1 70.44
81 0.29 L171 - E130 139 - 483 0 - 1 70.72
82 0.26 I207 - I120 173 - 473 0 - 1 70.99
83 0.23 R20 - T86 16 - 439 0 - 1 71.22
84 0.22 K233 - G272 199 - 625 0 - 1 71.44
85 0.22 R201 - E138 169 - 491 0 - 1 71.65
86 0.21 Q227 - T143 193 - 496 0 - 1 71.86
87 0.19 G226 - N119 192 - 472 0 - 1 72.05
88 0.17 R42 - A56 38 - 409 0 - 1 72.22
89 0.16 Q12 - T86 8 - 439 0 - 1 72.37
90 0.14 R42 - L55 38 - 408 0 - 1 72.51
91 0.14 L43 - W99 39 - 452 0 - 1 72.65
92 0.13 R232 - D246 198 - 599 0 - 1 72.78
93 0.12 R232 - C204 198 - 557 0 - 1 72.90
label freq
1 C237 5.90
2 Q19 5.15
3 Q227 4.13
4 K233 3.92
5 I26 3.76
6 I172 3.68
7 Q236 3.48
8 R228 2.98
9 F208 2.79
10 F222 2.63
11 W234 2.51
12 L30 2.49
13 Q170 2.39
14 R42 2.25
15 F238 2.00
16 N23 2.00
17 D33 1.96
18 E209 1.89
19 W281 1.86
20 A22 1.79
21 E16 1.34
22 R20 1.22
23 R201 1.22
24 G226 1.08
25 Y37 1.00
26 E27 1.00
27 D240 1.00
28 K34 0.99
29 N239 0.93
30 I207 0.90
31 R232 0.79
32 E15 0.61
33 V241 0.37
34 H220 0.30
35 L171 0.29
36 Q12 0.16
37 L43 0.14
label freq
1 L117 5.87
2 W99 5.30
3 K89 5.23
4 L55 3.71
5 N88 3.58
6 K57 2.99
7 Y145 2.96
8 W332 2.81
9 Y59 2.70
10 V90 2.51
11 D118 2.25
12 T86 2.12
13 N119 2.06
14 E130 1.94
15 G131 1.60
16 R68 1.52
17 D290 1.50
18 M101 1.49
19 R96 1.41
20 D246 1.13
21 T143 1.10
22 G162 1.00
23 D186 1.00
24 N230 1.00
25 K78 1.00
26 G53 1.00
27 Q75 1.00
28 A92 1.00
29 R314 0.98
30 N132 0.98
31 D83 0.97
32 D76 0.97
33 H91 0.96
34 S98 0.96
35 R134 0.94
36 G144 0.93
37 R129 0.90
38 D228 0.54
39 I120 0.26
40 G272 0.22
41 E138 0.22
42 A56 0.17
43 C204 0.12

The output
The following 93 contacts capture 72.90 (~98%) of the total frequency 74.50 (over 138 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 73 ctcs already capture 90.0% of 74.50.
The 73-th contact has a frequency of 0.57
let’s us know that ignoring frequencies < .1 reduces the full list from 138 to 93 entries while keeping ca 98% of \(\Sigma_t\). In this case, even if we have ctc_control=1.0 we don’t get 100% of the freqs reported (only 98%) because the min_freq=0.1 (default value) is causing the loss of %2 of contacts all of which have frequencies below 10%.
Interfaces: self_interface
From the docs of mdciao.cli.interface:
Note
----
If your definitions of `interface_selection_1` and
`interface_selection_2` lead to some overlap between
the interface members (see below), mdciao's default
is to ignore contact pairs within the same fragment.
E.g., in the context of a GPCR, computing
"TM3" vs "TM*" ("TM3" vs "all TMs") won't include
TM3-TM3 contacts by default. To include these
(or equivalent) contacts set `self_interface` = True.
Another example could be computing the interface of
C-terminus of a receptor with the entire receptor,
where it might be useful to include the contacts of
the C-terminus with itself.
When using `self_interface` = True, it's advisable to
increase `n_nearest`, since otherwise neighboring
residues of the shared set (the TM3-TM3 or the Cterm-Cterm)
will always appear as formed.
We can compute the self contacts of the $:nbsphinx-math:`alpha`$5-helix of the G-protein. Whereas most of the helix is straight, the C-terminal bends a bit backwards and interacts with itself:
[11]:
intf = mdciao.cli.interface("mdciao_example/traj.xtc", topology="mdciao_example/top.pdb",
fragments="consensus",accept_guess=True,
no_disk=True, interface_selection_1="G.H5", interface_selection_2="G.H5",
ctc_control=1.0,
GPCR_UniProt="adrb2_human",CGN_UniProt="gnas2_human",
self_interface=True,
#min_freq=0,
n_nearest=4,
figures=False)
intf.plot_freqs_as_bars(4.5, shorten_AAs=True, plot_atomtypes=True);
Will compute contact frequencies for trajectories:
mdciao_example/traj.xtc
with a stride of 1 frames
Using method 'resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 284 AAs GLU30 ( 760) - P0G395 (1043) (3) resSeq jumps
No local file ./adrb2_human.xlsx found, checking online in
https://gpcrdb.org/services/residues/extended/adrb2_human ...done!
Please cite the following reference to the GPCRdb:
* Kooistra et al, (2021) GPCRdb in 2021: Integrating GPCR sequence, structure and function
Nucleic Acids Research 49, D335--D343
https://doi.org/10.1093/nar/gkaa1080
For more information, call mdciao.nomenclature.references()
The GPCR-labels align best with fragments: [3] (first-last: GLU30-P0G395).
Mapping the GPCR fragments onto your topology:
TM1 with 32 AAs GLU30@1.29x29 ( 760) - PHE61@1.60x60 (791 ) (TM1)
ICL1 with 4 AAs GLU62@12.48x48 ( 792) - GLN65@12.51x51 (795 ) (ICL1)
TM2 with 32 AAs THR66@2.37x37 ( 796) - LYS97@2.68x67 (827 ) (TM2)
ECL1 with 4 AAs MET98@23.49x49 ( 828) - PHE101@23.52x52 (831 ) (ECL1)
TM3 with 36 AAs GLY102@3.21x21 ( 832) - SER137@3.56x56 (867 ) (TM3)
ICL2 with 8 AAs PRO138@34.50x50 ( 868) - LEU145@34.57x57 (875 ) (ICL2)
TM4 with 27 AAs THR146@4.38x38 ( 876) - HIS172@4.64x64 (902 ) (TM4)
ECL2 with 20 AAs TRP173 ( 903) - THR195 (922 ) (ECL2) resSeq jumps
TM5 with 42 AAs ASN196@5.35x36 ( 923) - GLU237@5.76x76 (964 ) (TM5)
ICL3 with 2 AAs GLY238 ( 965) - ARG239 (966 ) (ICL3)
TM6 with 35 AAs CYS265@6.27x27 ( 967) - GLN299@6.61x61 (1001) (TM6)
ECL3 with 4 AAs ASP300 (1002) - ILE303 (1005) (ECL3)
TM7 with 25 AAs ARG304@7.31x30 (1006) - ARG328@7.55x55 (1030) (TM7)
H8 with 12 AAs SER329@8.47x47 (1031) - LEU340@8.58x58 (1042) (H8)
No local file ./gnas2_human.xlsx found, checking online in
https://gpcrdb.org/services/residues/extended/gnas2_human ...done!
Please cite the following reference to the GPCRdb:
* Kooistra et al, (2021) GPCRdb in 2021: Integrating GPCR sequence, structure and function
Nucleic Acids Research 49, D335--D343
https://doi.org/10.1093/nar/gkaa1080
Please cite the following reference to the CGN nomenclature:
* Flock et al, (2015) Universal allosteric mechanism for G$\alpha$ activation by GPCRs
Nature 2015 524:7564 524, 173--179
https://doi.org/10.1038/nature14663
For more information, call mdciao.nomenclature.references()
The CGN-labels align best with fragments: [0] (first-last: LEU4-LEU394).
Mapping the CGN fragments onto your topology:
G.HN with 33 AAs LEU4@G.HN.10 ( 0) - VAL36@G.HN.53 (32 ) (G.HN)
G.hns1 with 3 AAs TYR37@G.hns1.01 ( 33) - ALA39@G.hns1.03 (35 ) (G.hns1)
G.S1 with 7 AAs THR40@G.S1.01 ( 36) - LEU46@G.S1.07 (42 ) (G.S1)
G.s1h1 with 6 AAs GLY47@G.s1h1.01 ( 43) - GLY52@G.s1h1.06 (48 ) (G.s1h1)
G.H1 with 7 AAs LYS53@G.H1.01 ( 49) - GLN59@G.H1.07 (55 ) (G.H1)
H.HA with 26 AAs LYS88@H.HA.04 ( 56) - LEU113@H.HA.29 (81 ) (H.HA)
H.hahb with 9 AAs VAL114@H.hahb.01 ( 82) - PRO122@H.hahb.09 (90 ) (H.hahb)
H.HB with 14 AAs GLU123@H.HB.01 ( 91) - ASN136@H.HB.14 (104 ) (H.HB)
H.hbhc with 7 AAs VAL137@H.hbhc.01 ( 105) - PRO143@H.hbhc.15 (111 ) (H.hbhc)
H.HC with 12 AAs PRO144@H.HC.01 ( 112) - GLU155@H.HC.12 (123 ) (H.HC)
H.hchd with 1 AAs ASP156@H.hchd.01 ( 124) - ASP156@H.hchd.01 (124 ) (H.hchd)
H.HD with 12 AAs GLU157@H.HD.01 ( 125) - GLU168@H.HD.12 (136 ) (H.HD)
H.hdhe with 5 AAs TYR169@H.hdhe.01 ( 137) - ASP173@H.hdhe.05 (141 ) (H.hdhe)
H.HE with 13 AAs CYS174@H.HE.01 ( 142) - LYS186@H.HE.13 (154 ) (H.HE)
H.hehf with 7 AAs GLN187@H.hehf.01 ( 155) - SER193@H.hehf.07 (161 ) (H.hehf)
H.HF with 6 AAs ASP194@H.HF.01 ( 162) - ARG199@H.HF.06 (167 ) (H.HF)
G.hfs2 with 5 AAs CYS200@G.hfs2.01 ( 168) - GLY206@G.hfs2.07 (172 ) (G.hfs2) resSeq jumps
G.S2 with 8 AAs ILE207@G.S2.01 ( 173) - VAL214@G.S2.08 (180 ) (G.S2)
G.s2s3 with 2 AAs ASP215@G.s2s3.01 ( 181) - LYS216@G.s2s3.02 (182 ) (G.s2s3)
G.S3 with 8 AAs VAL217@G.S3.01 ( 183) - VAL224@G.S3.08 (190 ) (G.S3)
G.s3h2 with 3 AAs GLY225@G.s3h2.01 ( 191) - GLN227@G.s3h2.03 (193 ) (G.s3h2)
G.H2 with 10 AAs ARG228@G.H2.01 ( 194) - CYS237@G.H2.10 (203 ) (G.H2)
G.h2s4 with 5 AAs PHE238@G.h2s4.01 ( 204) - THR242@G.h2s4.05 (208 ) (G.h2s4)
G.S4 with 7 AAs ALA243@G.S4.01 ( 209) - ALA249@G.S4.07 (215 ) (G.S4)
G.s4h3 with 8 AAs SER250@G.s4h3.01 ( 216) - ASN264@G.s4h3.15 (223 ) (G.s4h3) resSeq jumps
G.H3 with 18 AAs ARG265@G.H3.01 ( 224) - LEU282@G.H3.18 (241 ) (G.H3)
G.h3s5 with 3 AAs ARG283@G.h3s5.01 ( 242) - ILE285@G.h3s5.03 (244 ) (G.h3s5)
G.S5 with 7 AAs SER286@G.S5.01 ( 245) - ASN292@G.S5.07 (251 ) (G.S5)
G.s5hg with 1 AAs LYS293@G.s5hg.01 ( 252) - LYS293@G.s5hg.01 (252 ) (G.s5hg)
G.HG with 17 AAs GLN294@G.HG.01 ( 253) - ASP310@G.HG.17 (269 ) (G.HG)
G.hgh4 with 21 AAs TYR311@G.hgh4.01 ( 270) - ASP331@G.hgh4.21 (290 ) (G.hgh4)
G.H4 with 16 AAs PRO332@G.H4.01 ( 291) - ARG347@G.H4.17 (306 ) (G.H4)
G.h4s6 with 11 AAs ILE348@G.h4s6.01 ( 307) - TYR358@G.h4s6.20 (317 ) (G.h4s6)
G.S6 with 5 AAs CYS359@G.S6.01 ( 318) - PHE363@G.S6.05 (322 ) (G.S6)
G.s6h5 with 5 AAs THR364@G.s6h5.01 ( 323) - ASP368@G.s6h5.05 (327 ) (G.s6h5)
G.H5 with 26 AAs THR369@G.H5.01 ( 328) - LEU394@G.H5.26 (353 ) (G.H5)
Select group 1: G.H5
Select group 2: G.H5
Excluding contacts between 4 nearest neighbors reduces from 325 to 231 residue pairs. Use 'n_nearest' to control this (94 residue pairs discarded).
Will look for contacts in the interface between fragments
G.H5
and
G.H5.
Performing a first pass on the 231 group_1-group_2 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances.
Reduced to only 95 (from 231) residue pairs for the computation of actual residue-residue distances:
The following 4 contacts capture 3.97 (~100%) of the total frequency 3.99 (over 7 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 4 ctcs already capture 90.0% of 3.99.
The 4-th contact has a frequency of 0.97.
freq label residues fragments sum
1 1.00 R389@G.H5.21 - L394@G.H5.26 348 - 353 0 - 0 1.00
2 1.00 L388@G.H5.20 - L394@G.H5.26 347 - 353 0 - 0 2.00
3 1.00 L388@G.H5.20 - L393@G.H5.25 347 - 352 0 - 0 3.00
4 0.97 R385@G.H5.17 - L394@G.H5.26 344 - 353 0 - 0 3.97
label freq
1 L394@G.H5.26 2.97
2 L388@G.H5.20 2.00
3 R389@G.H5.21 1.00
4 L393@G.H5.25 1.00
5 R385@G.H5.17 0.97
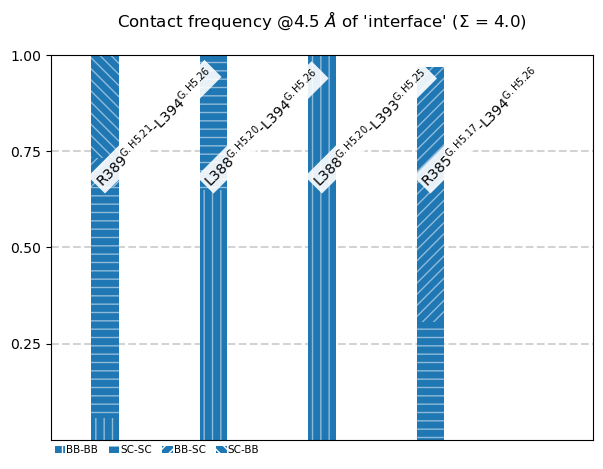
These are the contacts associated with the C-terminus of $:nbsphinx-math:`alpha`$5 bending back and interacting with itself:
freq label residues fragments sum
1 1.00 R389@G.H5.21 - L394@G.H5.26 348 - 353 0 - 0 1.00
2 1.00 L388@G.H5.20 - L394@G.H5.26 347 - 353 0 - 0 2.00
3 1.00 L388@G.H5.20 - L393@G.H5.25 347 - 352 0 - 0 3.00
4 0.97 R385@G.H5.17 - L394@G.H5.26 344 - 353 0 - 0 3.97
We have also ploted them also as bars, including the atom-types.
Interfaces: AA_selection
If the fragments themselves are still too broad a definition, one can select a sub-set of aminoacids of those fragments via AA_selection:
From the docs
AA_selection : str or list, default is None
Whatever the fragment definition and fragment selection
has been, one can further refine the list of
potential residue pairs by making a selection at
the level of single aminoacids (AAs).
E.g., if (like above) one has selected the interface
to be "TM3" vs "TM2",
>>> interface_selection_1="TM3"
>>> interface_selection_2="TM2"
but wants to select only some regions of those helices,
one can pass here an `AA_selection`.
Please read the rest of the docs, since the parameter has more options than the ones we’re about to use.
Here, we define the interface as contacts of the $:nbsphinx-math:`alpha`$5-helix of the G-protein with the TM-bundle, using
no_disk=True, interface_selection_1="G.H5", interface_selection_2="TM*"
and then use
AA_selection="390-394"
G.H5 with 26 AAs THR369@G.H5.01 ( 328) - LEU394@G.H5.26 (353 ) (G.H5)[12]:
intf = mdciao.cli.interface("mdciao_example/traj.xtc", topology="mdciao_example/top.pdb",
fragments="consensus",accept_guess=True,
no_disk=True, interface_selection_1="G.H5", interface_selection_2="TM*",
ctc_control=1.0,
GPCR_UniProt="adrb2_human",CGN_UniProt="gnas2_human",
n_nearest=4,
AA_selection="390-394",
figures=False)
intf.plot_freqs_as_bars(4.5, shorten_AAs=True, plot_atomtypes=True);
Will compute contact frequencies for trajectories:
mdciao_example/traj.xtc
with a stride of 1 frames
Using method 'resSeq+' these fragments were found
fragment 0 with 354 AAs LEU4 ( 0) - LEU394 (353 ) (0) resSeq jumps
fragment 1 with 340 AAs GLN1 ( 354) - ASN340 (693 ) (1)
fragment 2 with 66 AAs ALA2 ( 694) - PHE67 (759 ) (2)
fragment 3 with 284 AAs GLU30 ( 760) - P0G395 (1043) (3) resSeq jumps
No local file ./adrb2_human.xlsx found, checking online in
https://gpcrdb.org/services/residues/extended/adrb2_human ...done!
Please cite the following reference to the GPCRdb:
* Kooistra et al, (2021) GPCRdb in 2021: Integrating GPCR sequence, structure and function
Nucleic Acids Research 49, D335--D343
https://doi.org/10.1093/nar/gkaa1080
For more information, call mdciao.nomenclature.references()
The GPCR-labels align best with fragments: [3] (first-last: GLU30-P0G395).
Mapping the GPCR fragments onto your topology:
TM1 with 32 AAs GLU30@1.29x29 ( 760) - PHE61@1.60x60 (791 ) (TM1)
ICL1 with 4 AAs GLU62@12.48x48 ( 792) - GLN65@12.51x51 (795 ) (ICL1)
TM2 with 32 AAs THR66@2.37x37 ( 796) - LYS97@2.68x67 (827 ) (TM2)
ECL1 with 4 AAs MET98@23.49x49 ( 828) - PHE101@23.52x52 (831 ) (ECL1)
TM3 with 36 AAs GLY102@3.21x21 ( 832) - SER137@3.56x56 (867 ) (TM3)
ICL2 with 8 AAs PRO138@34.50x50 ( 868) - LEU145@34.57x57 (875 ) (ICL2)
TM4 with 27 AAs THR146@4.38x38 ( 876) - HIS172@4.64x64 (902 ) (TM4)
ECL2 with 20 AAs TRP173 ( 903) - THR195 (922 ) (ECL2) resSeq jumps
TM5 with 42 AAs ASN196@5.35x36 ( 923) - GLU237@5.76x76 (964 ) (TM5)
ICL3 with 2 AAs GLY238 ( 965) - ARG239 (966 ) (ICL3)
TM6 with 35 AAs CYS265@6.27x27 ( 967) - GLN299@6.61x61 (1001) (TM6)
ECL3 with 4 AAs ASP300 (1002) - ILE303 (1005) (ECL3)
TM7 with 25 AAs ARG304@7.31x30 (1006) - ARG328@7.55x55 (1030) (TM7)
H8 with 12 AAs SER329@8.47x47 (1031) - LEU340@8.58x58 (1042) (H8)
No local file ./gnas2_human.xlsx found, checking online in
https://gpcrdb.org/services/residues/extended/gnas2_human ...done!
Please cite the following reference to the GPCRdb:
* Kooistra et al, (2021) GPCRdb in 2021: Integrating GPCR sequence, structure and function
Nucleic Acids Research 49, D335--D343
https://doi.org/10.1093/nar/gkaa1080
Please cite the following reference to the CGN nomenclature:
* Flock et al, (2015) Universal allosteric mechanism for G$\alpha$ activation by GPCRs
Nature 2015 524:7564 524, 173--179
https://doi.org/10.1038/nature14663
For more information, call mdciao.nomenclature.references()
The CGN-labels align best with fragments: [0] (first-last: LEU4-LEU394).
Mapping the CGN fragments onto your topology:
G.HN with 33 AAs LEU4@G.HN.10 ( 0) - VAL36@G.HN.53 (32 ) (G.HN)
G.hns1 with 3 AAs TYR37@G.hns1.01 ( 33) - ALA39@G.hns1.03 (35 ) (G.hns1)
G.S1 with 7 AAs THR40@G.S1.01 ( 36) - LEU46@G.S1.07 (42 ) (G.S1)
G.s1h1 with 6 AAs GLY47@G.s1h1.01 ( 43) - GLY52@G.s1h1.06 (48 ) (G.s1h1)
G.H1 with 7 AAs LYS53@G.H1.01 ( 49) - GLN59@G.H1.07 (55 ) (G.H1)
H.HA with 26 AAs LYS88@H.HA.04 ( 56) - LEU113@H.HA.29 (81 ) (H.HA)
H.hahb with 9 AAs VAL114@H.hahb.01 ( 82) - PRO122@H.hahb.09 (90 ) (H.hahb)
H.HB with 14 AAs GLU123@H.HB.01 ( 91) - ASN136@H.HB.14 (104 ) (H.HB)
H.hbhc with 7 AAs VAL137@H.hbhc.01 ( 105) - PRO143@H.hbhc.15 (111 ) (H.hbhc)
H.HC with 12 AAs PRO144@H.HC.01 ( 112) - GLU155@H.HC.12 (123 ) (H.HC)
H.hchd with 1 AAs ASP156@H.hchd.01 ( 124) - ASP156@H.hchd.01 (124 ) (H.hchd)
H.HD with 12 AAs GLU157@H.HD.01 ( 125) - GLU168@H.HD.12 (136 ) (H.HD)
H.hdhe with 5 AAs TYR169@H.hdhe.01 ( 137) - ASP173@H.hdhe.05 (141 ) (H.hdhe)
H.HE with 13 AAs CYS174@H.HE.01 ( 142) - LYS186@H.HE.13 (154 ) (H.HE)
H.hehf with 7 AAs GLN187@H.hehf.01 ( 155) - SER193@H.hehf.07 (161 ) (H.hehf)
H.HF with 6 AAs ASP194@H.HF.01 ( 162) - ARG199@H.HF.06 (167 ) (H.HF)
G.hfs2 with 5 AAs CYS200@G.hfs2.01 ( 168) - GLY206@G.hfs2.07 (172 ) (G.hfs2) resSeq jumps
G.S2 with 8 AAs ILE207@G.S2.01 ( 173) - VAL214@G.S2.08 (180 ) (G.S2)
G.s2s3 with 2 AAs ASP215@G.s2s3.01 ( 181) - LYS216@G.s2s3.02 (182 ) (G.s2s3)
G.S3 with 8 AAs VAL217@G.S3.01 ( 183) - VAL224@G.S3.08 (190 ) (G.S3)
G.s3h2 with 3 AAs GLY225@G.s3h2.01 ( 191) - GLN227@G.s3h2.03 (193 ) (G.s3h2)
G.H2 with 10 AAs ARG228@G.H2.01 ( 194) - CYS237@G.H2.10 (203 ) (G.H2)
G.h2s4 with 5 AAs PHE238@G.h2s4.01 ( 204) - THR242@G.h2s4.05 (208 ) (G.h2s4)
G.S4 with 7 AAs ALA243@G.S4.01 ( 209) - ALA249@G.S4.07 (215 ) (G.S4)
G.s4h3 with 8 AAs SER250@G.s4h3.01 ( 216) - ASN264@G.s4h3.15 (223 ) (G.s4h3) resSeq jumps
G.H3 with 18 AAs ARG265@G.H3.01 ( 224) - LEU282@G.H3.18 (241 ) (G.H3)
G.h3s5 with 3 AAs ARG283@G.h3s5.01 ( 242) - ILE285@G.h3s5.03 (244 ) (G.h3s5)
G.S5 with 7 AAs SER286@G.S5.01 ( 245) - ASN292@G.S5.07 (251 ) (G.S5)
G.s5hg with 1 AAs LYS293@G.s5hg.01 ( 252) - LYS293@G.s5hg.01 (252 ) (G.s5hg)
G.HG with 17 AAs GLN294@G.HG.01 ( 253) - ASP310@G.HG.17 (269 ) (G.HG)
G.hgh4 with 21 AAs TYR311@G.hgh4.01 ( 270) - ASP331@G.hgh4.21 (290 ) (G.hgh4)
G.H4 with 16 AAs PRO332@G.H4.01 ( 291) - ARG347@G.H4.17 (306 ) (G.H4)
G.h4s6 with 11 AAs ILE348@G.h4s6.01 ( 307) - TYR358@G.h4s6.20 (317 ) (G.h4s6)
G.S6 with 5 AAs CYS359@G.S6.01 ( 318) - PHE363@G.S6.05 (322 ) (G.S6)
G.s6h5 with 5 AAs THR364@G.s6h5.01 ( 323) - ASP368@G.s6h5.05 (327 ) (G.s6h5)
G.H5 with 26 AAs THR369@G.H5.01 ( 328) - LEU394@G.H5.26 (353 ) (G.H5)
Select group 1: G.H5
Select group 2: TM1, TM2, TM3, TM4, TM5, TM6, TM7
Will look for contacts in the interface between fragments
G.H5
and
TM1, TM2, TM3, TM4, TM5, TM6, TM7.
Excluding residue pairs not involving residues '390-394' (5 AAs) reduces from 5954 to 1145 residue pairs.
Performing a first pass on the 1145 group_1-group_2 residue pairs to compute lower bounds on residue-residue distances via residue-COM distances.
Reduced to only 165 (from 1145) residue pairs for the computation of actual residue-residue distances:
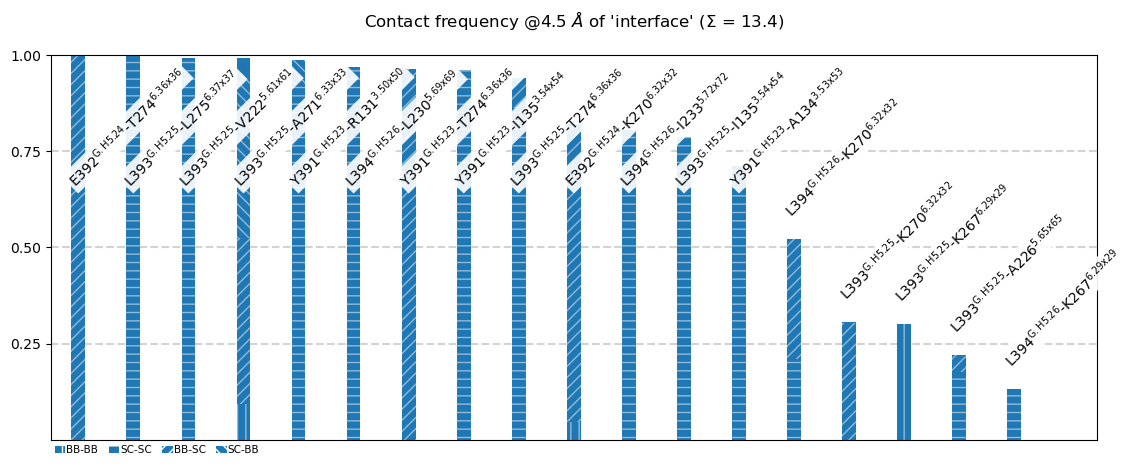
The following 18 contacts capture 13.40 (~98%) of the total frequency 13.64 (over 25 contacts with nonzero frequency at 4.50 Angstrom).
As orientation value, the first 14 ctcs already capture 90.0% of 13.64.
The 14-th contact has a frequency of 0.52.
freq label residues fragments sum
1 1.00 E392@G.H5.24 - T274@6.36x36 351 - 976 0 - 3 1.00
2 1.00 L393@G.H5.25 - L275@6.37x37 352 - 977 0 - 3 2.00
3 0.99 L393@G.H5.25 - V222@5.61x61 352 - 949 0 - 3 2.99
4 0.99 L393@G.H5.25 - A271@6.33x33 352 - 973 0 - 3 3.98
5 0.99 Y391@G.H5.23 - R131@3.50x50 350 - 861 0 - 3 4.97
6 0.97 L394@G.H5.26 - L230@5.69x69 353 - 957 0 - 3 5.94
7 0.96 Y391@G.H5.23 - T274@6.36x36 350 - 976 0 - 3 6.90
8 0.96 Y391@G.H5.23 - I135@3.54x54 350 - 865 0 - 3 7.86
9 0.94 L393@G.H5.25 - T274@6.36x36 352 - 976 0 - 3 8.80
10 0.82 E392@G.H5.24 - K270@6.32x32 351 - 972 0 - 3 9.62
11 0.80 L394@G.H5.26 - I233@5.72x72 353 - 960 0 - 3 10.42
12 0.79 L393@G.H5.25 - I135@3.54x54 352 - 865 0 - 3 11.21
13 0.71 Y391@G.H5.23 - A134@3.53x53 350 - 864 0 - 3 11.92
14 0.52 L394@G.H5.26 - K270@6.32x32 353 - 972 0 - 3 12.44
15 0.31 L393@G.H5.25 - K270@6.32x32 352 - 972 0 - 3 12.75
16 0.30 L393@G.H5.25 - K267@6.29x29 352 - 969 0 - 3 13.05
17 0.22 L393@G.H5.25 - A226@5.65x65 352 - 953 0 - 3 13.27
18 0.13 L394@G.H5.26 - K267@6.29x29 353 - 969 0 - 3 13.40
label freq
1 L393@G.H5.25 5.54
2 Y391@G.H5.23 3.62
3 L394@G.H5.26 2.42
4 E392@G.H5.24 1.82
label freq
1 T274@6.36x36 2.90
2 I135@3.54x54 1.75
3 K270@6.32x32 1.65
4 L275@6.37x37 1.00
5 V222@5.61x61 0.99
6 A271@6.33x33 0.99
7 R131@3.50x50 0.99
8 L230@5.69x69 0.97
9 I233@5.72x72 0.80
10 A134@3.53x53 0.71
11 K267@6.29x29 0.43
12 A226@5.65x65 0.22
Finally
Some of these parameters/criteria appear in other places in mdciao, not only at the moment of computing the distances, but also at the moment of showing them. E.g., the method mdciao.flare.freqs2flare automatically hides neighboring contacts via the exclude_neighbors = 1 parameter.
So, if at any moment you miss some contact in the reports (graphical or otherwise), check if some of the parameters above are at play.