Methods
The construction of the pathway is a two step process:
- construct a graph
- select the path of lowest energy
To construct the path, the root squared mean deviation (RMSD) is calculated between all pairs of conformation without superimposing them (i.e., using their given coordinates). If a chain is defined, only residues from that chain are used for the RMSD calculation. The RMSD for two molecules with atom positions x is defined as:
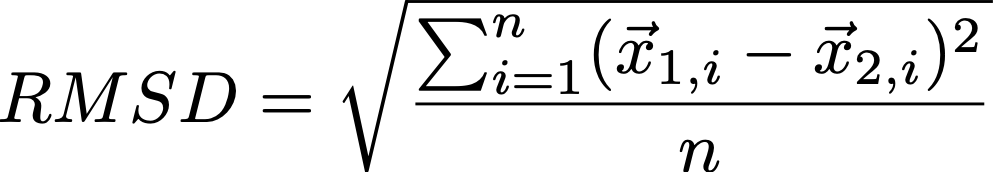
Defining a chain will increase the range of RMSD values. Consider the provided system in the example section. Here, the receptor undergoes only minor conformational changes. These atoms will contribute little to differences in RMSD between poses (conformations). Including them will lead to significantly lower RMSD values, due to the higher number of atoms in the formula. However, defining the chain is more a question of preference rather than having some actual rationale.
In the graph, each node represents a PDB file or molecular conformation. Starting from the first node, the path is constructed by connecting all nodes below the defined RMSD with an edge. Technically, the children are linked to their parents, allowing for a backtrace in the second step.
There are two criteria to select the lowest energy path:
- the lowest energy barrier
- for paths with the same barrier, the sum over shifted energies
The barrier is the highest energy along a given path. Since they are contained in all paths, the starting and final conformations are ignored for the barrier. The second criterion has two objectives. First, to favor shorter paths. Second, to achieve lower energies overall. To achieve both, the minimum enery is subtracted from all energies (no negative energies). The sum over the shifted energies fulfills both criteria.
The actual search for the path fulfilling both criteria is a modification of the dynamic programming approach, which results in a highly efficient backtrace.