Research
 |
 |
 |
| Analysis, prediction and visualization of molecular complexes | Structural dynamics of G protein coupled receptors (GPCRs) | Alzheimer's Disease |
|---|---|---|
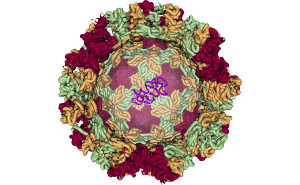
Our goal is to improve the understanding of proteins and their functions. Therefore we use knowledge based predictions of secon\ dary and tertiary structure interactions of helical membrane proteins. Furthermore, do we increase the explanatory power of protein models by fragment based modelling of proteins and protein complexes using Cryo-Electron Mi\ croscopy (EM) data as constraints. In order to share our findings with the research community we are developing web applications for analysis, prediction and visualization of proteins and\ their complexes. |
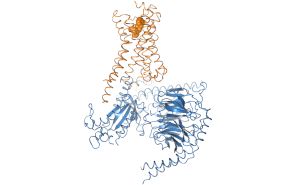
We work on the elucidation of the conformational space of GPCRs and its modulation by extracellular ligands and intracellular effector proteins by means\ of classical and umbrella sampling molecular dynamics simulations to address the issues of signaling specificity and biased signaling. For this reason we are computational modelling receptor G protein or arrestin interactions to elucidate the mechanism of signal transduction. |
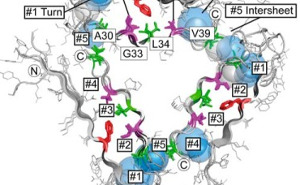
Influence of structural dynamics and oligomerisation of ϒ-secretase substrates (APP, APLP) on their being processed to cell toxic species. Structural modelling of the modulation of the ϒ-secretase section of APP by non-peptide active substances. Structural modelling of the modulation of fibril formation by peptide and non-peptide active substances. |
