D E T A I L S
What means high resolution?
The algorithm calculating volumes and packing densities is based on a grid. The default accuracy is corresponding to a grid size of 0.8 Å. High accuracy denotes a grid size of 0.4 Å. A further reduction of the grid size has only little benefit for the final results while dramatically increasing the calculation time.
Go to topWhat do the options for water mean?
Water molecules present in the uploaded PDB file can be handled in three different ways. Having waters present obviously will reduce number and/or size of cavities. Two of the options are obvious, keeping or erasing all water molecules in the PDB. The third option is to keep only inner waters. Waters are considered to be 'inner waters' if they are in holes that are not accessible for bulk water from the surrounding solution.
Go to topWhat do I see in the result section?
There are three different result views. All of them have four main elements displayed:
- Protein/RNA in a cartoon view, colored by packing density.
- Hetero atoms are shown as balls+sticks
- Side-chains forming cavities, colored by hydrophobicity.
- Cavities as spheres, colored by the averaged hydrophobicity of their constituing neighbors.
The first view that opens is an overview. You will see a small window with the molecule and the cavities. At the side there is an explanation of the coloring scheme. Below is a link to download all related files as ZIP archive. Furthermore, there is a link to the fullscreen view.
If you uploaded a ZIP archive containing multiple PDBs, you will see at the top of the molecule window the name of the current PDB and a link to the next, which will allow you to navigate through the results.
The fullscreen view opens first with a very simplified menu. There is a button to toggle all cavities. Another botton toggles all sidechains. The sidechains are displayed only if they constitute one of the cavities. Then there are two additional buttons for navigation. One leads back to the overview window, the other opens the full menu.
The full menu enables a lot of advanced selections and visualizations. Some of these are explained in these FAQs. More information can be found on NGL viewer manual.
What is the content of the download file?
There are three PDB formatted files, corresponding to the three main elements visualized by Voronoia. NAME.vor.pdb contains the uploaded PDB with the packing densities in the b-factor column.
NAME_neighbors.pdb contains only those residues from the previous file that form a cavity. The occupancy column contains the hydrophobicity of the residues.
NAME_holes.pdb contains the cavities as pseudo-atoms. The radius of the cavities is written into the b-factor and the averaged hydrophobicity of the neighboring residues into the occupancy.
Additionally, the file 'file_format.txt' explains the content of the PDB files in more detail. These four files are wrapped into a zip archive and provided for download.
Go to topHow to use the full NGL menu?
The NGL viewer provides a very powerful menu, that allows you a variety of modifications.
With the left mouse button clicked, the molecules can be rotated. With the right mouse botton clicked, the molecules can be shifted. With the mouse wheel one can zoom in and out.
Hovering over the molecule, information of the atoms is shown.
You can switch any element on/off by clicking on the 'eye'-symbol.

You can add representations by clicking the menu button on the top right of each section.

Then select under 'add' your preferred representation.
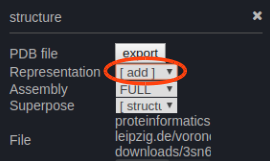
The filter symbol below a representation allows to select any group of atoms. More info in the manual.

More information can be found on NGL viewer manual.
Go to topHow to view results for all PDBs uploaded in a zip?
In the browser there are navigation links in the overview view of the result section. From the fullscreen views, one has to first return to the overview to access results of another PDB. On top of the molecule window the PDB-ID of the current molecules is shown and the ID of the next.
Go to topWhat is the content of 'browsePDB'?
It is a database of precalculated structures from the PDB, using the high-resolution mode and keeping internal waters. The database is updated on a weekly basis.
Go to topWhat if there is something missing in the DB?
The most simple solution is to download the PDB from e.g. rcsb.org and to upload it to Voronoia via the 'submit' form. If that is not a solution to you and you also don't want to wait for the next weekly update, drop us an email.
Go to topHow to navigate through different result views?
First, the overview opens. At its right bottom there is a link to the fullscreen view with the simple menu. From there you can either proceed to the full-menu view or back to the overview. Also from the full-menu view there is an option to return to the overview. In the fullscreen views the navigation buttons are located on the top left of the window.

How to create high resolution images?
To take pictures of the current window in the 'full-menu' view, select under 'File' in the top left either a quick snapshot or 'export image' with a few more options.
Generally the quality depends on the resolution of the cartoon representation. To adjust, open the menu in the cartoon section.

This will open the 'Representation' window. Increase the value of 'subdiv' to get smoother cartoons.

You may want to change the background color. On the top left, select 'View' and then 'Light theme'.
For further usage check full-menu view instructions.
Go to topHow to create a specific representation for a selection of atoms?
Either create a new representation or modify an existing one. To create a new representation, go the the menu on the top right in the 'molecules' section:

Then click 'add' representation:

and select e.g. 'surface'.
Once a new representation is created, filter the atoms that you want to highlight. Type e.g. 'hetero' next to the filter symbol. There is a entire atom selection language available: NGL selection. You can create as many representation and use the 'eye' symbol to switch them off/on.
Go to topHow to upload multiple pdbs?
The PDB files need to be wrapped into a .zip file. The zipped archive can then be uploaded either by clicking the 'Upload a PDB' banner or to drag & drop it onto the marked field.
Go to topAre there different kind of cavities?
There are four different kinds of cavities. The classification depends on both content and constituting neighbors.
One can easily navigate through them in the 'full-menu' view. In the 'cavities' section go to filter symbol. Enter either individual numbers or combinations, e.g. '1 or 2'.
Types of cavities:
- class '1': cavities with hetatm/water as direct neighbor
- class '2': empty cavities
- class '3': cavities with hetatm/waters as direct neighbor, but these are ignored
- class '4': cavities containing hetero atoms (no space for further waters)

Which hydrophocity scale is used?
Amino acid hydrophobicity scale from the supplementary information for "Recognition of transmembrane helices by the endoplasmic reticulum translocon," Hessa et al., Nature 433:377 (2005).
More negative means more hydrophobic. The met value is also assigned to selenomethionine.
| Amino acid | Hydrophobicity |
|---|---|
| asp | 3.49 |
| glu | 2.68 |
| asn | 2.05 |
| gln | 2.36 |
| lys | 2.71 |
| arg | 2.58 |
| his | 2.06 |
| gly | 0.74 |
| pro | 2.23 |
| ser | 0.84 |
| thr | 0.52 |
| cys | -0.13 |
| met | -0.10 |
| mse | -0.10 |
| ala | 0.11 |
| val | -0.31 |
| ile | -0.60 |
| leu | -0.55 |
| phe | -0.32 |
| trp | 0.30 |
| tyr | 0.68 |
Go to top